Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors
- PMID: 20418882
- PMCID: PMC2945309
- DOI: 10.1038/nsmb.1801
Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors
Abstract
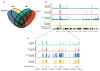
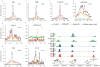
RNA polymerase (Pol) III transcribes many noncoding RNAs (for example, transfer RNAs) important for translational capacity and other functions. We localized Pol III, alternative TFIIIB complexes (BRF1 or BRF2) and TFIIIC in HeLa cells to determine the Pol III transcriptome, define gene classes and reveal 'TFIIIC-only' sites. Pol III localization in other transformed and primary cell lines reveals previously uncharacterized and cell type-specific Pol III loci as well as one microRNA. Notably, only a fraction of the in silico-predicted Pol III loci are occupied. Many occupied Pol III genes reside within an annotated Pol II promoter. Outside of Pol II promoters, occupied Pol III genes overlap with enhancer-like chromatin and enhancer-binding proteins such as ETS1 and STAT1. Moreover, Pol III occupancy scales with the levels of nearby Pol II, active chromatin and CpG content. These results suggest that active chromatin gates Pol III accessibility to the genome.
Figures




References
-
- White RJ. RNA Polymerase III Transcription. Springer-Verlag; New York, NY: 1998.
-
- Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23:614–22. - PubMed
-
- White RJ. RNA polymerases I and III, non-coding RNAs and cancer. Trends Genet. 2008;24:622–9. - PubMed
-
- Marshall L, White RJ. Non-coding RNA production by RNA polymerase III is implicated in cancer. Nat Rev Cancer. 2008;8:911–4. - PubMed
-
- Willis IM. RNA polymerase III. Genes, factors and transcriptional specificity. Eur J Biochem. 1993;212:1–11. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

