Microevolution of group A streptococci in vivo: capturing regulatory networks engaged in sociomicrobiology, niche adaptation, and hypervirulence
- PMID: 20418946
- PMCID: PMC2854683
- DOI: 10.1371/journal.pone.0009798
Microevolution of group A streptococci in vivo: capturing regulatory networks engaged in sociomicrobiology, niche adaptation, and hypervirulence
Abstract
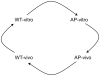
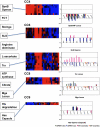
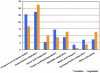
The onset of infection and the switch from primary to secondary niches are dramatic environmental changes that not only alter bacterial transcriptional programs, but also perturb their sociomicrobiology, often driving minor subpopulations with mutant phenotypes to prevail in specific niches. Having previously reported that M1T1 Streptococcus pyogenes become hypervirulent in mice due to selection of mutants in the covRS regulatory genes, we set out to dissect the impact of these mutations in vitro and in vivo from the impact of other adaptive events. Using a murine subcutaneous chamber model to sample the bacteria prior to selection or expansion of mutants, we compared gene expression dynamics of wild type (WT) and previously isolated animal-passaged (AP) covS mutant bacteria both in vitro and in vivo, and we found extensive transcriptional alterations of pathoadaptive and metabolic gene sets associated with invasion, immune evasion, tissue-dissemination, and metabolic reprogramming. In contrast to the virulence-associated differences between WT and AP bacteria, Phenotype Microarray analysis showed minor in vitro phenotypic differences between the two isogenic variants. Additionally, our results reflect that WT bacteria's rapid host-adaptive transcriptional reprogramming was not sufficient for their survival, and they were outnumbered by hypervirulent covS mutants with SpeB(-)/Sda(high) phenotype, which survived up to 14 days in mice chambers. Our findings demonstrate the engagement of unique regulatory modules in niche adaptation, implicate a critical role for bacterial genetic heterogeneity that surpasses transcriptional in vivo adaptation, and portray the dynamics underlying the selection of hypervirulent covS mutants over their parental WT cells.
Conflict of interest statement
Figures


References
-
- Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis. 2005;5:685–694. - PubMed
-
- Aziz RK. Saarbrücken, Germany: VDM Verlag; 2009. Molecular Dissection of the Clonal M1T1 Group A Streptococci.
-
- Kotb M, Norrby-Teglund A, McGeer A, El-Sherbini H, Dorak MT, et al. An immunogenetic and molecular basis for differences in outcomes of invasive group A streptococcal infections. Nat Med. 2002;8:1398–1404. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

