Specific recognition of influenza A/H1N1/2009 antibodies in human serum: a simple virus-free ELISA method
- PMID: 20418957
- PMCID: PMC2854694
- DOI: 10.1371/journal.pone.0010176
Specific recognition of influenza A/H1N1/2009 antibodies in human serum: a simple virus-free ELISA method
Abstract
Background: Although it has been estimated that pandemic Influenza A H1N1/2009 has infected millions of people from April to October 2009, a more precise figure requires a worldwide large-scale diagnosis of the presence of Influenza A/H1N1/2009 antibodies within the population. Assays typically used to estimate antibody titers (hemagglutination inhibition and microneutralization) would require the use of the virus, which would seriously limit broad implementation.
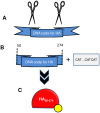
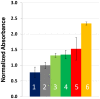
Methodology/principal findings: An ELISA method to evaluate the presence and relative concentration of specific Influenza A/H1N1/2009 antibodies in human serum samples is presented. The method is based on the use of a histidine-tagged recombinant fragment of the globular region of the hemagglutinin (HA) of the Influenza A H1N1/2009 virus expressed in E. coli.
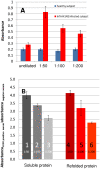
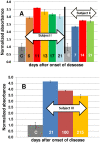
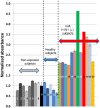
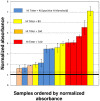
Conclusions/significance: The ELISA method consistently discerns between Inf A H1N1 infected and non-infected subjects, particularly after the third week of infection/exposure. Since it does not require the use of viral particles, it can be easily and quickly implemented in any basic laboratory. In addition, in a scenario of insufficient vaccine availability, the use of this ELISA could be useful to determine if a person has some level of specific antibodies against the virus and presumably at least partial protection.
Conflict of interest statement
Figures




References
-
- Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, et al. Mortality Associated With Influenza and Respiratory Syncytial Virus in the United States. JAMA. 2003;289 (2):179–1186. - PubMed
-
- Keech M, Beardsworth P. The impact of influenza on working days lost: A review of the literature. PharmacoEconomics. 2009;26 (11):911–924. - PubMed
-
- Molinari N-AM, Ortega-Sanchez IR, Messonnier ML, Thompson WW, Wortley PM, et al. The annual impact of seasonal influenza in the US: Measuring disease burden and costs. Vaccine. 2007;25 (27):5086–5096. - PubMed
-
- Carrat F, Flahault A. Influenza vaccine: The challenge of antigenic drift. Vaccine. 2007;25 (39–40):6852–6862. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

