Comparative genome analysis provides insights into the evolution and adaptation of Pseudomonas syringae pv. aesculi on Aesculus hippocastanum
- PMID: 20419105
- PMCID: PMC2856684
- DOI: 10.1371/journal.pone.0010224
Comparative genome analysis provides insights into the evolution and adaptation of Pseudomonas syringae pv. aesculi on Aesculus hippocastanum
Abstract
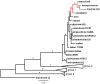
A recently emerging bleeding canker disease, caused by Pseudomonas syringae pathovar aesculi (Pae), is threatening European horse chestnut in northwest Europe. Very little is known about the origin and biology of this new disease. We used the nucleotide sequences of seven commonly used marker genes to investigate the phylogeny of three strains isolated recently from bleeding stem cankers on European horse chestnut in Britain (E-Pae). On the basis of these sequences alone, the E-Pae strains were identical to the Pae type-strain (I-Pae), isolated from leaf spots on Indian horse chestnut in India in 1969. The phylogenetic analyses also showed that Pae belongs to a distinct clade of P. syringae pathovars adapted to woody hosts. We generated genome-wide Illumina sequence data from the three E-Pae strains and one strain of I-Pae. Comparative genomic analyses revealed pathovar-specific genomic regions in Pae potentially implicated in virulence on a tree host, including genes for the catabolism of plant-derived aromatic compounds and enterobactin synthesis. Several gene clusters displayed intra-pathovar variation, including those encoding type IV secretion, a novel fatty acid biosynthesis pathway and a sucrose uptake pathway. Rates of single nucleotide polymorphisms in the four Pae genomes indicate that the three E-Pae strains diverged from each other much more recently than they diverged from I-Pae. The very low genetic diversity among the three geographically distinct E-Pae strains suggests that they originate from a single, recent introduction into Britain, thus highlighting the serious environmental risks posed by the spread of an exotic plant pathogenic bacterium to a new geographic location. The genomic regions in Pae that are absent from other P. syringae pathovars that infect herbaceous hosts may represent candidate genetic adaptations to infection of the woody parts of the tree.
Conflict of interest statement
Figures





References
-
- Strange RN, Scott PR. Plant disease: a threat to global food security. Annu Rev Phytopathol. 2005;43:83–116. - PubMed
-
- Brasier CM. The biosecurity threat to the UK and global environment from international trade in plants. Plant Pathol. 2008;57:792–808.
-
- Snyder LA, Loman N, Pallen MJ, Penn CW. Next-generation sequencing—the promise and perils of charting the great microbial unknown. Microb Ecol. 2009;57:1–3. - PubMed
-
- Webber JF, Parkinson NM, Rose J, Stanford H, Cook RTA, et al. Isolation and identification of Pseudomonas syringae pv. aesculi causing bleeding canker of horse chestnut in the UK. Plant Pathol New Dis Rep. 2008;15:1.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

