In vitro dissection of protein translocation into the mammalian endoplasmic reticulum
- PMID: 20419420
- PMCID: PMC3122127
- DOI: 10.1007/978-1-60327-412-8_20
In vitro dissection of protein translocation into the mammalian endoplasmic reticulum
Abstract
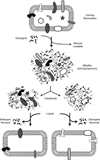
In eukaryotic cells, roughly one-fourth of all mRNAs code for secretory and membrane proteins. This class of proteins must first be segregated to the endoplasmic reticulum, where they are either translocated into the lumen or inserted into the lipid bilayer. The study of these processes has long relied on their successful reconstitution in cell-free systems. The high manipulability of such in vitro systems has allowed the identification of key machinery, elucidation of their functional roles in translocation, and dissection of their mechanisms of action. Here, we provide the basic methodology for (i) setting up robust mammalian-based in vitro translation and translocation systems, (ii) assays for protein translocation, insertion, and topology, and (iii) methods to solubilize, fractionate, and reconstitute ER membranes. Variations of these methods should be applicable not only to forward protein translocation systems but also for dissecting other poorly understood membrane-associated processes such as retrotranslocation.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

