BpaB, a novel protein encoded by the Lyme disease spirochete's cp32 prophages, binds to erp Operator 2 DNA
- PMID: 20421207
- PMCID: PMC2938228
- DOI: 10.1093/nar/gkq284
BpaB, a novel protein encoded by the Lyme disease spirochete's cp32 prophages, binds to erp Operator 2 DNA
Abstract
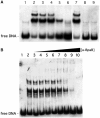
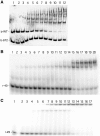
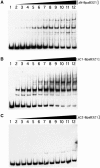
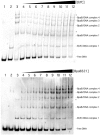
Borrelia burgdorferi produces Erp outer surface proteins throughout mammalian infection, but represses their synthesis during colonization of vector ticks. A DNA region 5' of the start of erp transcription, Operator 2, was previously shown to be essential for regulation of expression. We now report identification and characterization of a novel erp Operator 2-binding protein, which we named BpaB. erp operons are located on episomal cp32 prophages, and a single bacterium may contain as many as 10 different cp32s. Each cp32 family member encodes a unique BpaB protein, yet the three tested cp32-encoded BpaB alleles all bound to the same DNA sequence. A 20-bp region of erp Operator 2 was determined to be essential for BpaB binding, and initial protein binding to that site was required for binding of additional BpaB molecules. A 36-residue region near the BpaB carboxy terminus was found to be essential for high-affinity DNA-binding. BpaB competed for binding to erp Operator 2 with a second B. burgdorferi DNA-binding protein, EbfC. Thus, cellular levels of free BpaB and EbfC could potentially control erp transcription levels.
Figures
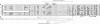

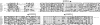


Similar articles
-
Borrelia burgdorferi cp32 BpaB modulates expression of the prophage NucP nuclease and SsbP single-stranded DNA-binding protein.J Bacteriol. 2012 Sep;194(17):4570-8. doi: 10.1128/JB.00661-12. Epub 2012 Jun 22. J Bacteriol. 2012. PMID: 22730122 Free PMC article.
-
Erp and Rev Adhesins of the Lyme Disease Spirochete's Ubiquitous cp32 Prophages Assist the Bacterium during Vertebrate Infection.Infect Immun. 2023 Mar 15;91(3):e0025022. doi: 10.1128/iai.00250-22. Epub 2023 Feb 28. Infect Immun. 2023. PMID: 36853019 Free PMC article. Review.
-
Borrelia burgdorferi EbfC, a novel, chromosomally encoded protein, binds specific DNA sequences adjacent to erp loci on the spirochete's resident cp32 prophages.J Bacteriol. 2006 Jun;188(12):4331-9. doi: 10.1128/JB.00005-06. J Bacteriol. 2006. PMID: 16740939 Free PMC article.
-
BpaB and EbfC DNA-binding proteins regulate production of the Lyme disease spirochete's infection-associated Erp surface proteins.J Bacteriol. 2012 Feb;194(4):778-86. doi: 10.1128/JB.06394-11. Epub 2011 Dec 9. J Bacteriol. 2012. PMID: 22155777 Free PMC article.
-
Repetition, conservation, and variation: the multiple cp32 plasmids of Borrelia species.J Mol Microbiol Biotechnol. 2000 Oct;2(4):411-22. J Mol Microbiol Biotechnol. 2000. PMID: 11075913 Review.
Cited by
-
Gene Regulation and Transcriptomics.Curr Issues Mol Biol. 2021;42:223-266. doi: 10.21775/cimb.042.223. Epub 2020 Dec 10. Curr Issues Mol Biol. 2021. PMID: 33300497 Free PMC article.
-
Borrelia burgdorferi cp32 BpaB modulates expression of the prophage NucP nuclease and SsbP single-stranded DNA-binding protein.J Bacteriol. 2012 Sep;194(17):4570-8. doi: 10.1128/JB.00661-12. Epub 2012 Jun 22. J Bacteriol. 2012. PMID: 22730122 Free PMC article.
-
Gac Is a Transcriptional Repressor of the Lyme Disease Spirochete's OspC Virulence-Associated Surface Protein.J Bacteriol. 2023 Apr 25;205(4):e0044022. doi: 10.1128/jb.00440-22. Epub 2023 Mar 15. J Bacteriol. 2023. PMID: 36920207 Free PMC article.
-
Erp and Rev Adhesins of the Lyme Disease Spirochete's Ubiquitous cp32 Prophages Assist the Bacterium during Vertebrate Infection.Infect Immun. 2023 Mar 15;91(3):e0025022. doi: 10.1128/iai.00250-22. Epub 2023 Feb 28. Infect Immun. 2023. PMID: 36853019 Free PMC article. Review.
-
Peaceful coexistence amongst Borrelia plasmids: getting by with a little help from their friends?Plasmid. 2013 Sep;70(2):161-7. doi: 10.1016/j.plasmid.2013.05.002. Epub 2013 May 30. Plasmid. 2013. PMID: 23727020 Free PMC article. Review.
References
-
- Stanek G, Strle F. Lyme borreliosis. Lancet. 2003;362:1639–1647. - PubMed
-
- Steere AC. Lyme disease. New Engl. J. Med. 2001;345:115–125. - PubMed
-
- Stevenson B, Zückert WR, Akins DR. In: The Spirochetes: Molecular and Cellular Biology. Saier MH, García-Lara J, editors. Oxford: Horizon Press; 2001. pp. 87–100.
-
- Stevenson B, Bykowski T, Cooley AE, Babb K, Miller JC, Woodman ME, von Lackum K, Riley SP. In: Molecular Biology of Spirochetes. Cabello FC, Godfrey HP, Hulinska D, editors. Amsterdam: IOS Press; 2006. pp. 354–372.

