An extended set of PRDM1/BLIMP1 target genes links binding motif type to dynamic repression
- PMID: 20421211
- PMCID: PMC2938208
- DOI: 10.1093/nar/gkq268
An extended set of PRDM1/BLIMP1 target genes links binding motif type to dynamic repression
Abstract
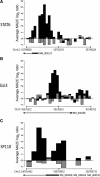
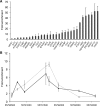
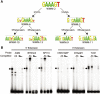
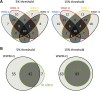
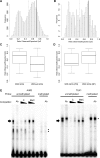
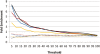
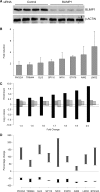
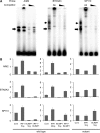
The transcriptional repressor B lymphocyte-induced maturation protein-1 (BLIMP1) regulates gene expression and cell fate. The DNA motif bound by BLIMP1 in vitro overlaps with that of interferon regulatory factors (IRFs), which respond to inflammatory/immune signals. At such sites, BLIMP1 and IRFs can antagonistically regulate promoter activity. In vitro motif selection predicts that only a subset of BLIMP1 or IRF sites is subject to antagonistic regulation, but the extent to which antagonism occurs is unknown, since an unbiased assessment of BLIMP1 occupancy in vivo is lacking. To address this, we identified an extended set of promoters occupied by BLIMP1. Motif discovery and enrichment analysis demonstrate that multiple motif variants are required to capture BLIMP1 binding specificity. These are differentially associated with CpG content, leading to the observation that BLIMP1 DNA-binding is methylation sensitive. In occupied promoters, only a subset of BLIMP1 motifs overlap with IRF motifs. Conversely, a distinct subset of IRF motifs is not enriched amongst occupied promoters. Genes linked to occupied promoters containing overlapping BLIMP1/IRF motifs (e.g. AIM2, SP110, BTN3A3) are shown to constitute a dynamic target set which is preferentially activated by BLIMP1 knock-down. These data confirm and extend the competitive model of BLIMP1 and IRF interaction.
Figures
Similar articles
-
Repression of IFN-gamma induction of class II transactivator: a role for PRDM1/Blimp-1 in regulation of cytokine signaling.J Immunol. 2006 Oct 1;177(7):4584-93. doi: 10.4049/jimmunol.177.7.4584. J Immunol. 2006. PMID: 16982896
-
Stage-specific modulation of IFN-regulatory factor 4 function by Krüppel-type zinc finger proteins.J Immunol. 2001 May 15;166(10):6104-11. doi: 10.4049/jimmunol.166.10.6104. J Immunol. 2001. PMID: 11342629
-
B lymphocyte-induced maturation protein (Blimp)-1, IFN regulatory factor (IRF)-1, and IRF-2 can bind to the same regulatory sites.J Immunol. 2004 Nov 1;173(9):5556-63. doi: 10.4049/jimmunol.173.9.5556. J Immunol. 2004. PMID: 15494505
-
Direct Inhibition of IRF-Dependent Transcriptional Regulatory Mechanisms Associated With Disease.Front Immunol. 2019 May 24;10:1176. doi: 10.3389/fimmu.2019.01176. eCollection 2019. Front Immunol. 2019. PMID: 31178872 Free PMC article. Review.
-
Conserved and Unique Functions of Blimp1 in Immune Cells.Front Immunol. 2022 Jan 27;12:805260. doi: 10.3389/fimmu.2021.805260. eCollection 2021. Front Immunol. 2022. PMID: 35154079 Free PMC article. Review.
Cited by
-
Aiolos collaborates with Blimp-1 to regulate the survival of multiple myeloma cells.Cell Death Differ. 2016 Jul;23(7):1175-84. doi: 10.1038/cdd.2015.167. Epub 2016 Jan 29. Cell Death Differ. 2016. PMID: 26823144 Free PMC article.
-
Integrative multiomics reveals common endotypes across PSEN1, PSEN2, and APP mutations in familial Alzheimer's disease.Alzheimers Res Ther. 2025 Jan 4;17(1):5. doi: 10.1186/s13195-024-01659-6. Alzheimers Res Ther. 2025. PMID: 39754192 Free PMC article.
-
SPIB and BATF provide alternate determinants of IRF4 occupancy in diffuse large B-cell lymphoma linked to disease heterogeneity.Nucleic Acids Res. 2014 Jul;42(12):7591-610. doi: 10.1093/nar/gku451. Epub 2014 May 29. Nucleic Acids Res. 2014. PMID: 24875472 Free PMC article.
-
Tau Pathology Drives Dementia Risk-Associated Gene Networks toward Chronic Inflammatory States and Immunosuppression.Cell Rep. 2020 Nov 17;33(7):108398. doi: 10.1016/j.celrep.2020.108398. Cell Rep. 2020. PMID: 33207193 Free PMC article.
-
PRDM1 controls the sequential activation of neural, neural crest and sensory progenitor determinants.Development. 2019 Dec 16;146(24):dev181107. doi: 10.1242/dev.181107. Development. 2019. PMID: 31806661 Free PMC article.
References
-
- Bikoff EK, Morgan MA, Robertson EJ. An expanding job description for Blimp-1/PRDM1. Curr. Opin. Genet. Dev. 2009;19:379–385. - PubMed
-
- Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19:607–620. - PubMed
-
- Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, Giltnane JM, Yang L, Zhao H, Calame K, et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. - PubMed
-
- Kallies A, Hawkins ED, Belz GT, Metcalf D, Hommel M, Corcoran LM, Hodgkin PD, Nutt SL. Transcriptional repressor Blimp-1 is essential for T cell homeostasis and self-tolerance. Nat. Immunol. 2006;7:466–474. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

