Myogenic microRNA expression requires ATP-dependent chromatin remodeling enzyme function
- PMID: 20421421
- PMCID: PMC2897572
- DOI: 10.1128/MCB.00214-10
Myogenic microRNA expression requires ATP-dependent chromatin remodeling enzyme function
Abstract
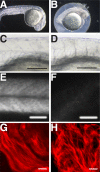
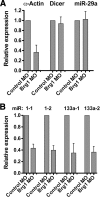
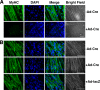
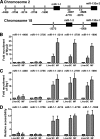
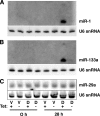
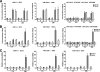
Knockdown of the Brg1 ATPase subunit of SWI/SNF chromatin remodeling enzymes in developing zebrafish caused stunted tail formation and altered sarcomeric actin organization, which phenocopies the loss of the microRNA processing enzyme Dicer, or the knockdown of myogenic microRNAs. Furthermore, myogenic microRNA expression and differentiation was blocked in Brg1 conditional myoblasts differentiated ex vivo. The binding of Brg1 upstream of myogenic microRNA sequences correlated with MyoD binding and accessible chromatin structure in satellite cells and myofibers, and it was required for chromatin accessibility and microRNA expression in a tissue culture model for myogenesis. The results implicate ATP-dependent chromatin remodelers in myogenic microRNA gene regulation.
Figures




References
-
- Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281-297. - PubMed
-
- Berkes, C. A., D. A. Bergstrom, B. H. Penn, K. J. Seaver, P. S. Knoepfler, and S. J. Tapscott. 2004. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell 14:465-477. - PubMed
-
- Bernstein, E., S. Y. Kim, M. A. Carmell, E. P. Murchison, H. Alcorn, M. Z. Li, A. A. Mills, S. J. Elledge, K. V. Anderson, and G. J. Hannon. 2003. Dicer is essential for mouse development. Nat. Genet. 35:215-217. - PubMed
-
- Bultman, S., T. Gebuhr, D. Yee, C. La Mantia, J. Nicholson, A. Gilliam, F. Randazzo, D. Metzger, P. Chambon, G. Crabtree, and T. Magnuson. 2000. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 6:1287-1295. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
