Pathway discovery in mantle cell lymphoma by integrated analysis of high-resolution gene expression and copy number profiling
- PMID: 20421449
- PMCID: PMC2924229
- DOI: 10.1182/blood-2010-01-263806
Pathway discovery in mantle cell lymphoma by integrated analysis of high-resolution gene expression and copy number profiling
Abstract
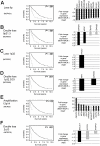
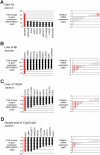
The genome of mantle cell lymphoma (MCL) is, in addition to the translocation t(11;14), characterized by a high number of secondary chromosomal gains and losses that probably account for the various survival times of MCL patients. We investigated 77 primary MCL tumors with available clinical information using high-resolution RNA expression and genomic profiling and applied our recently developed gene expression and dosage integrator algorithm to identify novel genes and pathways that may be of relevance for the pathobiology of MCL. We show that copy number neutral loss of heterozygosity is common in MCL and targets regions that are frequently affected by deletions. The molecular consequences of genomic copy number changes appear complex, even in genomic loci with identified tumor suppressors, such as the region 9p21 containing the CDKN2A locus. Moreover, the deregulation of novel genes, such as CUL4A, ING1, and MCPH1, may affect the 2 crucial pathogenetic mechanisms in MCL, the disturbance of the proliferation, and DNA damage response pathways. Deregulation of the Hippo pathway may have a pathogenetic role in MCL because decreased expression of its members MOBKL2A, MOBKL2B, and LATS2 was associated with inferior outcome, including an independent validation series of 32 MCLs.
Figures



Comment in
-
Discovery of Hippo in MCL.Blood. 2010 Aug 12;116(6):861-2. doi: 10.1182/blood-2010-05-284075. Blood. 2010. PMID: 20705764 No abstract available.
References
-
- Swerdlow SH, Campo E, Harris NL, et al., editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th ed. Lyon, France: IARC; 2008.
-
- Jares P, Colomer D, Campo E. Genetic and molecular pathogenesis of mantle cell lymphoma: perspectives for new targeted therapeutics. Nat Rev Cancer. 2007;7(10):750–762. - PubMed
-
- Rosenwald A, Wright G, Wiestner A, et al. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 2003;3(2):185–197. - PubMed
-
- Orchard J, Garand R, Davis Z, et al. A subset of t(11;14) lymphoma with mantle cell features displays mutated IgVH genes and includes patients with good prognosis, nonnodal disease. Blood. 2003;101(12):4975–4981. - PubMed
-
- Nodit L, Bahler DW, Jacobs SA, Locker J, Swerdlow SH. Indolent mantle cell lymphoma with nodal involvement and mutated immunoglobulin heavy chain genes. Hum Pathol. 2003;34(10):1030–1034. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

