Surprising fitness consequences of GC-biased gene conversion: I. Mutation load and inbreeding depression
- PMID: 20421602
- PMCID: PMC2907210
- DOI: 10.1534/genetics.110.116368
Surprising fitness consequences of GC-biased gene conversion: I. Mutation load and inbreeding depression
Erratum in
- Genetics. 2012 Apr;190(4):1585
Abstract
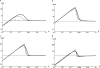
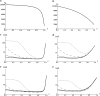
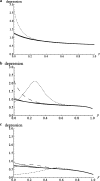
GC-biased gene conversion (gBGC) is a recombination-associated process mimicking selection in favor of G and C alleles. It is increasingly recognized as a widespread force in shaping the genomic nucleotide landscape. In recombination hotspots, gBGC can lead to bursts of fixation of GC nucleotides and to accelerated nucleotide substitution rates. It was recently shown that these episodes of strong gBGC could give spurious signatures of adaptation and/or relaxed selection. There is also evidence that gBGC could drive the fixation of deleterious amino acid mutations in some primate genes. This raises the question of the potential fitness effects of gBGC. While gBGC has been metaphorically termed the "Achilles' heel" of our genome, we do not know whether interference between gBGC and selection merely has practical consequences for the analysis of sequence data or whether it has broader fundamental implications for individuals and populations. I developed a population genetics model to predict the consequences of gBGC on the mutation load and inbreeding depression. I also used estimates available for humans to quantitatively evaluate the fitness impact of gBGC. Surprising features emerged from this model: (i) Contrary to classical mutation load models, gBGC generates a fixation load independent of population size and could contribute to a significant part of the load; (ii) gBGC can maintain recessive deleterious mutations for a long time at intermediate frequency, in a similar way to overdominance, and these mutations generate high inbreeding depression, even if they are slightly deleterious; (iii) since mating systems affect both the selection efficacy and gBGC intensity, gBGC challenges classical predictions concerning the interaction between mating systems and deleterious mutations, and gBGC could constitute an additional cost of outcrossing; and (iv) if mutations are biased toward A and T alleles, very low gBGC levels can reduce the load. A robust prediction is that the gBGC level minimizing the load depends only on the mutational bias and population size. These surprising results suggest that gBGC may have nonnegligible fitness consequences and could play a significant role in the evolution of genetic systems. They also shed light on the evolution of gBGC itself.
Figures


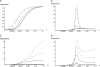



References
-
- Abramowitz, M., and I. A. Stegun, 1970. Handbook of Mathematical Functions. Dover, New York.
-
- Bataillon, T., and M. Kirkpatrick, 2000. Inbreeding depression due to mildly deleterious mutations in finite populations: size does matter. Genet. Res. 75 75–81. - PubMed
-
- Bengtsson, B. O., 1990. The effect of biased conversion on the mutation load. Genet. Res. 55 183–187. - PubMed
-
- Bengtsson, B. O., and M. K. Uyenoyama, 1990. Evolution of the segregation ratio: modification of gene conversion and meiotic drive. Theor. Popul. Biol. 38 192–218. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

