A fluorescence in situ hybridization system for karyotyping soybean
- PMID: 20421607
- PMCID: PMC2907198
- DOI: 10.1534/genetics.109.113753
A fluorescence in situ hybridization system for karyotyping soybean
Abstract
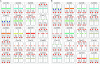
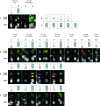
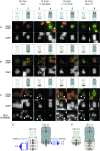
The development of a universal soybean (Glycine max [L.] Merr.) cytogenetic map that associates classical genetic linkage groups, molecular linkage groups, and a sequence-based physical map with the karyotype has been impeded due to the soybean chromosomes themselves, which are small and morphologically homogeneous. To overcome this obstacle, we screened soybean repetitive DNA to develop a cocktail of fluorescent in situ hybridization (FISH) probes that could differentially label mitotic chromosomes in root tip preparations. We used genetically anchored BAC clones both to identify individual chromosomes in metaphase spreads and to complete a FISH-based karyotyping cocktail that permitted simultaneous identification of all 20 chromosome pairs. We applied these karyotyping tools to wild soybean, G. soja Sieb. and Zucc., which represents a large gene pool of potentially agronomically valuable traits. These studies led to the identification and characterization of a reciprocal chromosome translocation between chromosomes 11 and 13 in two accessions of wild soybean. The data confirm that this translocation is widespread in G. soja accessions and likely accounts for the semi-sterility found in some G. soja by G. max crosses.
Figures





References
-
- Ahmad, Q. N., E. J. Britten and D. E. Byth, 1983. A quantitative method of karyotypic analysis applied to the soybean, Glycine max. Cytologia 48 879–892.
-
- Ahmad, Q. N., E. J. Britten and D. E. Byth, 1984. The karyotype of Glycine soja and its relationship to that of the soybean, Glycine max. Cytologia 49 645–658.
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215 403–410. - PubMed
-
- Arumuganathan, K., and E. D. Earle, 1991. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9 208–218.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

