A microarray-based genetic screen for yeast chronological aging factors
- PMID: 20421943
- PMCID: PMC2858703
- DOI: 10.1371/journal.pgen.1000921
A microarray-based genetic screen for yeast chronological aging factors
Abstract
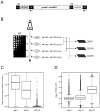
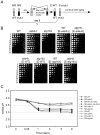
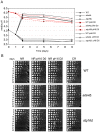
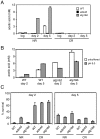
Model organisms have played an important role in the elucidation of multiple genes and cellular processes that regulate aging. In this study we utilized the budding yeast, Saccharomyces cerevisiae, in a large-scale screen for genes that function in the regulation of chronological lifespan, which is defined by the number of days that non-dividing cells remain viable. A pooled collection of viable haploid gene deletion mutants, each tagged with unique identifying DNA "bar-code" sequences was chronologically aged in liquid culture. Viable mutants in the aging population were selected at several time points and then detected using a microarray DNA hybridization technique that quantifies abundance of the barcode tags. Multiple short- and long-lived mutants were identified using this approach. Among the confirmed short-lived mutants were those defective for autophagy, indicating a key requirement for the recycling of cellular organelles in longevity. Defects in autophagy also prevented lifespan extension induced by limitation of amino acids in the growth media. Among the confirmed long-lived mutants were those defective in the highly conserved de novo purine biosynthesis pathway (the ADE genes), which ultimately produces IMP and AMP. Blocking this pathway extended lifespan to the same degree as calorie (glucose) restriction. A recently discovered cell-extrinsic mechanism of chronological aging involving acetic acid secretion and toxicity was suppressed in a long-lived ade4Delta mutant and exacerbated by a short-lived atg16Delta autophagy mutant. The identification of multiple novel effectors of yeast chronological lifespan will greatly aid in the elucidation of mechanisms that cells and organisms utilize in slowing down the aging process.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Gene-nutrient interaction markedly influences yeast chronological lifespan.Exp Gerontol. 2016 Dec 15;86:113-123. doi: 10.1016/j.exger.2016.04.012. Epub 2016 Apr 25. Exp Gerontol. 2016. PMID: 27125759 Free PMC article.
-
Genome-wide screen in Saccharomyces cerevisiae identifies vacuolar protein sorting, autophagy, biosynthetic, and tRNA methylation genes involved in life span regulation.PLoS Genet. 2010 Jul 15;6(7):e1001024. doi: 10.1371/journal.pgen.1001024. PLoS Genet. 2010. PMID: 20657825 Free PMC article.
-
Chronological aging-induced apoptosis in yeast.Biochim Biophys Acta. 2008 Jul;1783(7):1280-5. doi: 10.1016/j.bbamcr.2008.03.017. Epub 2008 Apr 10. Biochim Biophys Acta. 2008. PMID: 18445486 Free PMC article. Review.
-
Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae.Aging Cell. 2009 Aug;8(4):353-69. doi: 10.1111/j.1474-9726.2009.00469.x. Epub 2009 Apr 21. Aging Cell. 2009. PMID: 19302372 Free PMC article.
-
Amino acid homeostasis and chronological longevity in Saccharomyces cerevisiae.Subcell Biochem. 2012;57:161-86. doi: 10.1007/978-94-007-2561-4_8. Subcell Biochem. 2012. PMID: 22094422 Free PMC article. Review.
Cited by
-
Gene-nutrient interaction markedly influences yeast chronological lifespan.Exp Gerontol. 2016 Dec 15;86:113-123. doi: 10.1016/j.exger.2016.04.012. Epub 2016 Apr 25. Exp Gerontol. 2016. PMID: 27125759 Free PMC article.
-
Interplays of AMPK and TOR in Autophagy Regulation in Yeast.Cells. 2023 Feb 4;12(4):519. doi: 10.3390/cells12040519. Cells. 2023. PMID: 36831186 Free PMC article. Review.
-
Role of autophagy in aging: The good, the bad, and the ugly.Aging Cell. 2023 Jan;22(1):e13753. doi: 10.1111/acel.13753. Epub 2022 Dec 20. Aging Cell. 2023. PMID: 36539927 Free PMC article. Review.
-
Uncovering Natural Longevity Alleles from Intercrossed Pools of Aging Fission Yeast Cells.Genetics. 2018 Oct;210(2):733-744. doi: 10.1534/genetics.118.301262. Epub 2018 Aug 2. Genetics. 2018. PMID: 30072377 Free PMC article.
-
Potential benefits of medium chain fatty acids in aging and neurodegenerative disease.Front Aging Neurosci. 2023 Aug 23;15:1230467. doi: 10.3389/fnagi.2023.1230467. eCollection 2023. Front Aging Neurosci. 2023. PMID: 37680538 Free PMC article. Review.
References
-
- Partridge L. Some highlights of research on aging with invertebrates, 2008. Aging Cell. 2008;7:605–608. - PubMed
-
- Hansen M, Hsu AL, Dillin A, Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005;1:e17. doi: 10.1371/journal.pgen.0010017. - DOI - PMC - PubMed
-
- Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183:1751–1752. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

