In vitro reconstitution of SARS-coronavirus mRNA cap methylation
- PMID: 20421945
- PMCID: PMC2858705
- DOI: 10.1371/journal.ppat.1000863
In vitro reconstitution of SARS-coronavirus mRNA cap methylation
Erratum in
- PLoS Pathog. 2010;6(5). doi: 10.1371/annotation/a0dde376-2eb1-4ce3-8887-d29f5ba6f162
Abstract
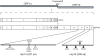
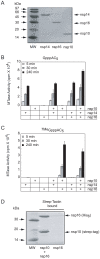
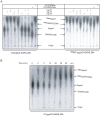
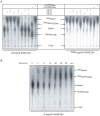
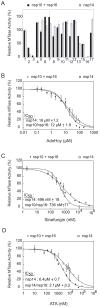
SARS-coronavirus (SARS-CoV) genome expression depends on the synthesis of a set of mRNAs, which presumably are capped at their 5' end and direct the synthesis of all viral proteins in the infected cell. Sixteen viral non-structural proteins (nsp1 to nsp16) constitute an unusually large replicase complex, which includes two methyltransferases putatively involved in viral mRNA cap formation. The S-adenosyl-L-methionine (AdoMet)-dependent (guanine-N7)-methyltransferase (N7-MTase) activity was recently attributed to nsp14, whereas nsp16 has been predicted to be the AdoMet-dependent (nucleoside-2'O)-methyltransferase. Here, we have reconstituted complete SARS-CoV mRNA cap methylation in vitro. We show that mRNA cap methylation requires a third viral protein, nsp10, which acts as an essential trigger to complete RNA cap-1 formation. The obligate sequence of methylation events is initiated by nsp14, which first methylates capped RNA transcripts to generate cap-0 (7Me)GpppA-RNAs. The latter are then selectively 2'O-methylated by the 2'O-MTase nsp16 in complex with its activator nsp10 to give rise to cap-1 (7Me)GpppA(2'OMe)-RNAs. Furthermore, sensitive in vitro inhibition assays of both activities show that aurintricarboxylic acid, active in SARS-CoV infected cells, targets both MTases with IC(50) values in the micromolar range, providing a validated basis for anti-coronavirus drug design.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2'O)-methyltransferase activity.J Virol. 2008 Aug;82(16):8071-84. doi: 10.1128/JVI.00407-08. Epub 2008 Apr 16. J Virol. 2008. PMID: 18417574 Free PMC article.
-
Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex.PLoS Pathog. 2011 Oct;7(10):e1002294. doi: 10.1371/journal.ppat.1002294. Epub 2011 Oct 13. PLoS Pathog. 2011. PMID: 22022266 Free PMC article.
-
Biochemical characterization of the (nucleoside-2'O)-methyltransferase activity of dengue virus protein NS5 using purified capped RNA oligonucleotides (7Me)GpppAC(n) and GpppAC(n).J Gen Virol. 2010 Jan;91(Pt 1):112-21. doi: 10.1099/vir.0.015511-0. Epub 2009 Sep 23. J Gen Virol. 2010. PMID: 19776234
-
Coronavirus non-structural protein 16: evasion, attenuation, and possible treatments.Virus Res. 2014 Dec 19;194:191-9. doi: 10.1016/j.virusres.2014.09.009. Epub 2014 Sep 30. Virus Res. 2014. PMID: 25278144 Free PMC article. Review.
-
Coronavirus 2'-O-methyltransferase: A promising therapeutic target.Virus Res. 2023 Oct 15;336:199211. doi: 10.1016/j.virusres.2023.199211. Epub 2023 Sep 4. Virus Res. 2023. PMID: 37634741 Free PMC article. Review.
Cited by
-
[Replication and transmission mechanisms of highly pathogenic human coronavirus].Zhejiang Da Xue Xue Bao Yi Xue Ban. 2020 May 25;49(3):324-339. doi: 10.3785/j.issn.1008-9292.2020.03.16. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2020. PMID: 32762165 Free PMC article. Review. Chinese.
-
Bisubstrate Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 Nsp14 Methyltransferase.ACS Med Chem Lett. 2022 Jul 22;13(9):1477-1484. doi: 10.1021/acsmedchemlett.2c00265. eCollection 2022 Sep 8. ACS Med Chem Lett. 2022. PMID: 36097498 Free PMC article.
-
Structure of SARS-CoV-2 MTase nsp14 with the inhibitor STM957 reveals inhibition mechanism that is shared with a poxviral MTase VP39.J Struct Biol X. 2024 Jul 29;10:100109. doi: 10.1016/j.yjsbx.2024.100109. eCollection 2024 Dec. J Struct Biol X. 2024. PMID: 39188530 Free PMC article.
-
mRNA maturation in giant viruses: variation on a theme.Nucleic Acids Res. 2015 Apr 20;43(7):3776-88. doi: 10.1093/nar/gkv224. Epub 2015 Mar 16. Nucleic Acids Res. 2015. PMID: 25779049 Free PMC article.
-
The molecular virology of coronaviruses.J Biol Chem. 2020 Sep 11;295(37):12910-12934. doi: 10.1074/jbc.REV120.013930. Epub 2020 Jul 13. J Biol Chem. 2020. PMID: 32661197 Free PMC article. Review.
References
-
- Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. - PubMed
-
- Thiel V, Ivanov KA, Putics A, Hertzig T, Schelle B, et al. Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol. 2003;84:2305–2315. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

