FAM-MDR: a flexible family-based multifactor dimensionality reduction technique to detect epistasis using related individuals
- PMID: 20421984
- PMCID: PMC2858665
- DOI: 10.1371/journal.pone.0010304
FAM-MDR: a flexible family-based multifactor dimensionality reduction technique to detect epistasis using related individuals
Abstract
We propose a novel multifactor dimensionality reduction method for epistasis detection in small or extended pedigrees, FAM-MDR. It combines features of the Genome-wide Rapid Association using Mixed Model And Regression approach (GRAMMAR) with Model-Based MDR (MB-MDR). We focus on continuous traits, although the method is general and can be used for outcomes of any type, including binary and censored traits. When comparing FAM-MDR with Pedigree-based Generalized MDR (PGMDR), which is a generalization of Multifactor Dimensionality Reduction (MDR) to continuous traits and related individuals, FAM-MDR was found to outperform PGMDR in terms of power, in most of the considered simulated scenarios. Additional simulations revealed that PGMDR does not appropriately deal with multiple testing and consequently gives rise to overly optimistic results. FAM-MDR adequately deals with multiple testing in epistasis screens and is in contrast rather conservative, by construction. Furthermore, simulations show that correcting for lower order (main) effects is of utmost importance when claiming epistasis. As Type 2 Diabetes Mellitus (T2DM) is a complex phenotype likely influenced by gene-gene interactions, we applied FAM-MDR to examine data on glucose area-under-the-curve (GAUC), an endophenotype of T2DM for which multiple independent genetic associations have been observed, in the Amish Family Diabetes Study (AFDS). This application reveals that FAM-MDR makes more efficient use of the available data than PGMDR and can deal with multi-generational pedigrees more easily. In conclusion, we have validated FAM-MDR and compared it to PGMDR, the current state-of-the-art MDR method for family data, using both simulations and a practical dataset. FAM-MDR is found to outperform PGMDR in that it handles the multiple testing issue more correctly, has increased power, and efficiently uses all available information.
Conflict of interest statement
Figures

 ,
,  and
and  , for the analysis without main effects correction.
, for the analysis without main effects correction.
 and
and  . Analyses are performed without correction for main effects. The first three panels show results of FAM-MDR (A), FAM-MDR* (B) and PGMDR (C) for the usual situation study considering 10 SNPs. The final panel (D) shows PGMDR results when only 2 SNPs are considered. The straight lines indicate the theoretical probability-probability curve (light blue) and the 5% significance level (dark blue).
. Analyses are performed without correction for main effects. The first three panels show results of FAM-MDR (A), FAM-MDR* (B) and PGMDR (C) for the usual situation study considering 10 SNPs. The final panel (D) shows PGMDR results when only 2 SNPs are considered. The straight lines indicate the theoretical probability-probability curve (light blue) and the 5% significance level (dark blue).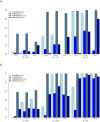
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

