Evaluation of the psoriasis transcriptome across different studies by gene set enrichment analysis (GSEA)
- PMID: 20422035
- PMCID: PMC2857878
- DOI: 10.1371/journal.pone.0010247
Evaluation of the psoriasis transcriptome across different studies by gene set enrichment analysis (GSEA)
Abstract
Background: Our objective was to develop a consistent molecular definition of psoriasis. There have been several published microarray studies of psoriasis, and we compared disease-related genes identified across these different studies of psoriasis with our own in order to establish a consensus.
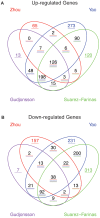
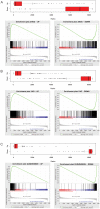
Methodology/principal findings: We present a psoriasis transcriptome from a group of 15 patients enrolled in a clinical study, and assessed its biological validity using a set of important pathways known to be involved in psoriasis. We also identified a key set of cytokines that are now strongly implicated in driving disease-related pathology, but which are not detected well on gene array platforms and require more sensitive methods to measure mRNA levels in skin tissues. Comparison of our transcriptome with three other published lists of psoriasis genes showed apparent inconsistencies based on the number of overlapping genes. We extended the well-established approach of Gene Set Enrichment Analysis (GSEA) to compare a new study with these other published list of differentially expressed genes (DEG) in a more comprehensive manner. We applied our method to these three published psoriasis transcriptomes and found them to be in good agreement with our study.
Conclusions/significance: Due to wide variability in clinical protocols, platform and sample handling, and subtle disease-related signals, intersection of published DEG lists was unable to establish consensus between studies. In order to leverage the power of multiple transcriptomes reported by several laboratories using different patients and protocols, more sophisticated methods like the extension of GSEA presented here, should be used in order to overcome the shortcomings of overlapping individual DEG approach.
Conflict of interest statement
Figures
Similar articles
-
Cross-study homogeneity of psoriasis gene expression in skin across a large expression range.PLoS One. 2013;8(1):e52242. doi: 10.1371/journal.pone.0052242. Epub 2013 Jan 4. PLoS One. 2013. PMID: 23308107 Free PMC article. Clinical Trial.
-
Anti-TNF- αtreatment-related pathways and biomarkers revealed by transcriptome analysis in Chinese psoriasis patients.BMC Syst Biol. 2019 Apr 5;13(Suppl 2):29. doi: 10.1186/s12918-019-0698-7. BMC Syst Biol. 2019. PMID: 30953507 Free PMC article.
-
Meta-analysis derived (MAD) transcriptome of psoriasis defines the "core" pathogenesis of disease.PLoS One. 2012;7(9):e44274. doi: 10.1371/journal.pone.0044274. Epub 2012 Sep 5. PLoS One. 2012. PMID: 22957057 Free PMC article.
-
The treatment of psoriasis with etanercept.Semin Cutan Med Surg. 2005 Mar;24(1):28-36. doi: 10.1016/j.sder.2005.01.003. Semin Cutan Med Surg. 2005. PMID: 15900796 Review.
-
Etanercept: an overview of its role in the treatment of psoriasis.Expert Opin Drug Metab Toxicol. 2008 Mar;4(3):305-10. doi: 10.1517/17425255.4.3.305. Expert Opin Drug Metab Toxicol. 2008. PMID: 18363545 Review.
Cited by
-
Cross-study homogeneity of psoriasis gene expression in skin across a large expression range.PLoS One. 2013;8(1):e52242. doi: 10.1371/journal.pone.0052242. Epub 2013 Jan 4. PLoS One. 2013. PMID: 23308107 Free PMC article. Clinical Trial.
-
Progressive activation of T(H)2/T(H)22 cytokines and selective epidermal proteins characterizes acute and chronic atopic dermatitis.J Allergy Clin Immunol. 2012 Dec;130(6):1344-54. doi: 10.1016/j.jaci.2012.07.012. Epub 2012 Aug 27. J Allergy Clin Immunol. 2012. PMID: 22951056 Free PMC article.
-
Suppression of molecular inflammatory pathways by Toll-like receptor 7, 8, and 9 antagonists in a model of IL-23-induced skin inflammation.PLoS One. 2013 Dec 27;8(12):e84634. doi: 10.1371/journal.pone.0084634. eCollection 2013. PLoS One. 2013. PMID: 24386404 Free PMC article.
-
Exploration of Prognostic Immune-Related Genes and lncRNAs Biomarkers in Kidney Renal Clear Cell Carcinoma and Its Crosstalk with Acute Kidney Injury.J Oncol. 2022 Feb 8;2022:6100187. doi: 10.1155/2022/6100187. eCollection 2022. J Oncol. 2022. PMID: 35178091 Free PMC article.
-
Prognostic value of nicotinamide adenine dinucleotide (NAD+) metabolic genes in patients with stomach adenocarcinoma based on bioinformatics analysis.J Gastrointest Oncol. 2022 Dec;13(6):2845-2862. doi: 10.21037/jgo-22-1092. J Gastrointest Oncol. 2022. PMID: 36636067 Free PMC article.
References
-
- Irizarry RA, Warren D, Spencer F, Kim IF, Biswal S, et al. Multiple-laboratory comparison of microarray platforms. Nat Methods. 2005;2:345–350. - PubMed
-
- Larkin JE, Frank BC, Gavras H, Sultana R, Quackenbush J. Independence and reproducibility across microarray platforms. Nat Methods. 2005;2:337–343. - PubMed
-
- Shi LM, Perkins RG, Fang H, Tong WD. Reproducible and reliable microarray results through quality control: good laboratory proficiency and appropriate data analysis practices are essential. Current Opinion in Biotechnology. 2008;19:10–18. - PubMed
-
- Suarez-Farinas M, Magnasco MO. Comparing microarray studies. Methods Mol Biol. 2007;377:139–152. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

