Mass spectrometric imaging for biomedical tissue analysis
- PMID: 20423155
- PMCID: PMC2907483
- DOI: 10.1021/cr100012c
Mass spectrometric imaging for biomedical tissue analysis
Figures
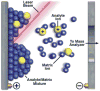
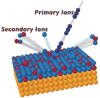

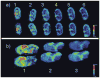

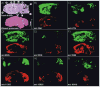
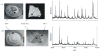
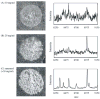
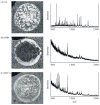






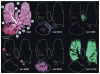


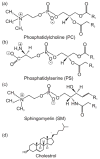
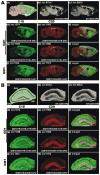

References
-
- Siuzdak G. The Expanding Role of Mass Spectrometry in Biotechnology. MCC Press; San Diego, CA: 2006. p. 11.
-
- Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Science. 1989;246:64. - PubMed
-
- Karas M, Bachmann D, Hillenkamp F. Anal Chem. 1985;57:2935.
-
- Caprioli RM, Farmer TB, Gile J. Anal Chem. 1997;69:4751. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

