EST analysis reveals putative genes involved in glycyrrhizin biosynthesis
- PMID: 20423525
- PMCID: PMC2886062
- DOI: 10.1186/1471-2164-11-268
EST analysis reveals putative genes involved in glycyrrhizin biosynthesis
Abstract
Background: Glycyrrhiza uralensis is one of the most popular medicinal plants in the world and is also widely used in the flavoring of food and tobacco. Due to limited genomic and transcriptomic data, the biosynthetic pathway of glycyrrhizin, the major bioactive compound in G. uralensis, is currently unclear. Identification of candidate genes involved in the glycyrrhizin biosynthetic pathway will significantly contribute to the understanding of the biosynthetic and medicinal chemistry of this compound.
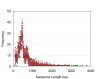
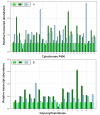
Results: We used the 454 GS FLX platform and Titanium regents to produce a substantial expressed sequence tag (EST) dataset from the vegetative organs of G. uralensis. A total of 59,219 ESTs with an average read length of 409 bp were generated. 454 ESTs were combined with the 50,666 G. uralensis ESTs in GenBank. The combined ESTs were assembled into 27,229 unique sequences (11,694 contigs and 15,535 singletons). A total of 20,437 unique gene elements representing approximately 10,000 independent transcripts were annotated using BLAST searches (e-value <or= 1e-5) against the SwissProt, KEGG, TAIR, Nr and Nt databases. The assembled sequences were annotated with gene names and Gene Ontology (GO) terms. With respect to the genes related to glycyrrhizin metabolism, genes encoding 16 enzymes of the 18 total steps of the glycyrrhizin skeleton synthesis pathway were found. To identify novel genes that encode cytochrome P450 enzymes and glycosyltransferases, which are related to glycyrrhizin metabolism, a total of 125 and 172 unigenes were found to be homologous to cytochrome P450s and glycosyltransferases, respectively. The cytochrome P450 candidate genes were classified into 32 CYP families, while the glycosyltransferase candidate genes were classified into 45 categories by GO analysis. Finally, 3 cytochrome P450 enzymes and 6 glycosyltransferases were selected as the candidates most likely to be involved in glycyrrhizin biosynthesis through an organ-specific expression pattern analysis based on real-time PCR.
Conclusions: Using the 454 GS FLX platform and Titanium reagents, our study provides a high-quality EST database for G. uralensis. Based on the EST analysis, novel candidate genes related to the secondary metabolite pathway of glycyrrhizin, including novel genes encoding cytochrome P450s and glycosyltransferases, were found. With the assistance of organ-specific expression pattern analysis, 3 unigenes encoding cytochrome P450s and 6 unigenes encoding glycosyltransferases were selected as the candidates most likely to be involved in glycyrrhizin biosynthesis.
Figures



References
-
- Parkinson J. Expressed Sequence Tags (ESTs) Generation and Analysis. Humana Press; 2009.
-
- Brautigam A, Shrestha RP, Whitten D, Wilkerson CG, Carr KM, Froehlich JE, Weber AP. Low-coverage massively parallel pyrosequencing of cDNAs enables proteomics in non-model species: comparison of a species-specific database generated by pyrosequencing with databases from related species for proteome analysis of pea chloroplast envelopes. J Biotechnol. 2008;136(1-2):44–53. doi: 10.1016/j.jbiotec.2008.02.007. - DOI - PubMed
-
- Shibata S. A drug over the millennia: pharmacognosy, chemistry, and pharmacology of licorice. Yakugaku Zasshi. 2000;120(10):849–862. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

