CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer
- PMID: 20423907
- PMCID: PMC2938198
- DOI: 10.1093/nar/gkq285
CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer
Abstract
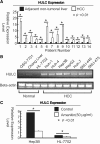
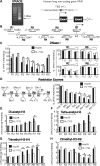
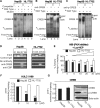
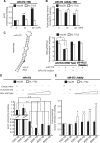
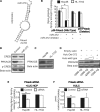
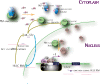
Long non-coding RNA (lncRNA), highly up-regulated in liver cancer (HULC) plays an important role in tumorigenesis. Depletion of HULC resulted in a significant deregulation of several genes involved in liver cancer. Although up-regulation of HULC expression in hepatocellular carcinoma has been reported, the molecular mechanisms remain unknown. In this study, we used in vivo and in vitro approaches to characterize cancer-dependent alterations in the chromatin organization and find a CREB binding site (encompassing from -67 to -53 nt) in the core promoter. Besides, we also provided evidence that PKA pathway may involved in up-regulation of HULC. Furthermore, we demonstrated HULC may act as an endogenous 'sponge', which down-regulates a series of microRNAs (miRNAs) activities, including miR-372. Inhibition of miR-372 leads to reducing translational repression of its target gene, PRKACB, which in turn induces phosphorylation of CREB. Over-expression of miR-372 decreases the association of CREB with the proximal promoter, followed by the dissociation of P300, resulting in a change of the histone 'code', such as in deacetylation and methylation. The study elucidates that fine tuning of HULC expression is part of an auto-regulatory loop in which it's inhibitory to expression and activity of miR-372 allows lncRNA up-regulated expression in liver cancer.
Figures




References
-
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long nonocoding RNAs. Cell. 2009;136:629–641. - PubMed
-
- Wang J, Zhang J, Zheng H, Li J, Liu D, Li H, Samudrala R, Yu J, Wong GK. Mouse transcriptome: neutral evolution of ‘non-coding’ complementary DNAs. Nature. 2004;431:757–761. - PubMed
-
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. - PubMed
-
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. - PubMed
-
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

