Genomic organization of eukaryotic tRNAs
- PMID: 20426822
- PMCID: PMC2888827
- DOI: 10.1186/1471-2164-11-270
Genomic organization of eukaryotic tRNAs
Abstract
Background: Surprisingly little is known about the organization and distribution of tRNA genes and tRNA-related sequences on a genome-wide scale. While tRNA gene complements are usually reported in passing as part of genome annotation efforts, and peculiar features such as the tandem arrangements of tRNA gene in Entamoeba histolytica have been described in some detail, systematic comparative studies are rare and mostly restricted to bacteria. We therefore set out to survey the genomic arrangement of tRNA genes and pseudogenes in a wide range of eukaryotes to identify common patterns and taxon-specific peculiarities.
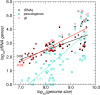
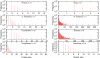
Results: In line with previous reports, we find that tRNA complements evolve rapidly and tRNA gene and pseudogene locations are subject to rapid turnover. At phylum level, the distributions of the number of tRNA genes and pseudogenes numbers are very broad, with standard deviations on the order of the mean. Even among closely related species we observe dramatic changes in local organization. For instance, 65% and 87% of the tRNA genes and pseudogenes are located in genomic clusters in zebrafish and stickleback, resp., while such arrangements are relatively rare in the other three sequenced teleost fish genomes. Among basal metazoa, Trichoplax adherens has hardly any duplicated tRNA gene, while the sea anemone Nematostella vectensis boasts more than 17000 tRNA genes and pseudogenes. Dramatic variations are observed even within the eutherian mammals. Higher primates, for instance, have 616 +/- 120 tRNA genes and pseudogenes of which 17% to 36% are arranged in clusters, while the genome of the bushbaby Otolemur garnetti has 45225 tRNA genes and pseudogenes of which only 5.6% appear in clusters. In contrast, the distribution is surprisingly uniform across plant genomes. Consistent with this variability, syntenic conservation of tRNA genes and pseudogenes is also poor in general, with turn-over rates comparable to those of unconstrained sequence elements. Despite this large variation in abundance in Eukarya we observe a significant correlation between the number of tRNA genes, tRNA pseudogenes, and genome size.
Conclusions: The genomic organization of tRNA genes and pseudogenes shows complex lineage-specific patterns characterized by an extensive variability that is in striking contrast to the extreme levels of sequence-conservation of the tRNAs themselves. The comprehensive analysis of the genomic organization of tRNA genes and pseudogenes in Eukarya provides a basis for further studies into the interplay of tRNA gene arrangements and genome organization in general.
Figures


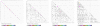


References
-
- Gesteland RF, Atkins JF, Eds. The RNA World. Plainview, NY: Cold Spring Harbor Laboratory Press; 1993.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

