Consolidating metabolite identifiers to enable contextual and multi-platform metabolomics data analysis
- PMID: 20426876
- PMCID: PMC2879285
- DOI: 10.1186/1471-2105-11-214
Consolidating metabolite identifiers to enable contextual and multi-platform metabolomics data analysis
Abstract
Background: Analysis of data from high-throughput experiments depends on the availability of well-structured data that describe the assayed biomolecules. Procedures for obtaining and organizing such meta-data on genes, transcripts and proteins have been streamlined in many data analysis packages, but are still lacking for metabolites. Chemical identifiers are notoriously incoherent, encompassing a wide range of different referencing schemes with varying scope and coverage. Online chemical databases use multiple types of identifiers in parallel but lack a common primary key for reliable database consolidation. Connecting identifiers of analytes found in experimental data with the identifiers of their parent metabolites in public databases can therefore be very laborious.
Results: Here we present a strategy and a software tool for integrating metabolite identifiers from local reference libraries and public databases that do not depend on a single common primary identifier. The program constructs groups of interconnected identifiers of analytes and metabolites to obtain a local metabolite-centric SQLite database. The created database can be used to map in-house identifiers and synonyms to external resources such as the KEGG database. New identifiers can be imported and directly integrated with existing data. Queries can be performed in a flexible way, both from the command line and from the statistical programming environment R, to obtain data set tailored identifier mappings.
Conclusions: Efficient cross-referencing of metabolite identifiers is a key technology for metabolomics data analysis. We provide a practical and flexible solution to this task and an open-source program, the metabolite masking tool (MetMask), available at http://metmask.sourceforge.net, that implements our ideas.
Figures

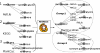
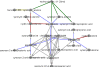

References
-
- Tokimatsu T, Sakurai N, Suzuki H, Ohta H, Nishitani K, Koyama T, Umezawa T, Misawa N, Saito K, Shibata D. KaPPA-view: a web-based analysis tool for integration of transcript and metabolite data on plant metabolic pathway maps. Plant Physiol. 2005;138(3):1289–1300. doi: 10.1104/pp.105.060525. - DOI - PMC - PubMed
-
- Usadel B, Nagel A, Thimm O, Redestig H, Bläsing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D, Scheible WR, Gibon Y, Morcuende R, Weicht D, Meyer S, Stitt M. Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol. 2005;138(3):1195–1204. doi: 10.1104/pp.105.060459. - DOI - PMC - PubMed
-
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–73. doi: 10.1038/ng1180. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

