Translational repression by PUF proteins in vitro
- PMID: 20427513
- PMCID: PMC2874173
- DOI: 10.1261/rna.2070110
Translational repression by PUF proteins in vitro
Abstract
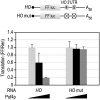
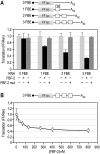
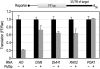
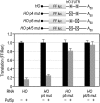
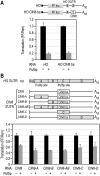
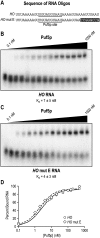
PUF (Pumilio and FBF) proteins provide a paradigm for mRNA regulatory proteins. They interact with specific sequences in the 3' untranslated regions (UTRs) of target mRNAs and cause changes in RNA stability or translational activity. Here we describe an in vitro translation assay that reconstitutes the translational repression activity of canonical PUF proteins. In this system, recombinant PUF proteins were added to yeast cell lysates to repress reporter mRNAs bearing the 3'UTRs of specific target mRNAs. PUF proteins from Saccharomyces cerevisiae and Caenorhabditis elegans were active in the assay and were specific by multiple criteria. Puf5p, a yeast PUF protein, repressed translation of four target RNAs. Repression mediated by the HO 3'UTR was particularly efficient, due to a specific sequence in that 3'UTR. The sequence lies downstream from the PUF binding site and does not affect PUF protein binding. PUF-mediated repression was sensitive to the distance between the ORF and the regulatory elements in the 3'UTR: excessive distance decreased repression activity. Our data demonstrate that PUF proteins function in vitro across species, that different mRNA targets are regulated differentially, and that specific ancillary sequences distinguish one yeast mRNA target from another. We suggest a model in which PUF proteins can control translation termination or elongation.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
