Spindly/CCDC99 is required for efficient chromosome congression and mitotic checkpoint regulation
- PMID: 20427577
- PMCID: PMC2883941
- DOI: 10.1091/mbc.e09-04-0356
Spindly/CCDC99 is required for efficient chromosome congression and mitotic checkpoint regulation
Abstract
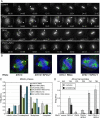
Spindly recruits a fraction of cytoplasmic dynein to kinetochores for poleward movement of chromosomes and control of mitotic checkpoint signaling. Here we show that human Spindly is a cell cycle-regulated mitotic phosphoprotein that interacts with the Rod/ZW10/Zwilch (RZZ) complex. The kinetochore levels of Spindly are regulated by microtubule attachment and biorientation induced tension. Deletion mutants lacking the N-terminal half of the protein (NDelta253), or the conserved Spindly box (DeltaSB), strongly localized to kinetochores and failed to respond to attachment or tension. In addition, these mutants prevented the removal of the RZZ complex and that of MAD2 from bioriented chromosomes and caused cells to arrest at metaphase, showing that RZZ-Spindly has to be removed from kinetochores to terminate mitotic checkpoint signaling. Depletion of Spindly by RNAi, however, caused cells to arrest in prometaphase because of a delay in microtubule attachment. Surprisingly, this defect was alleviated by codepletion of ZW10. Thus, Spindly is not only required for kinetochore localization of dynein but is a functional component of a mechanism that couples dynein-dependent poleward movement of chromosomes to their efficient attachment to microtubules.
Figures






References
-
- Ashar H. R., James L., Gray K., Carr D., Black S., Armstrong L., Bishop W. R., Kirschmeier P. Farnesyl transferase inhibitors block the farnesylation of CENP-E and CENP-F and alter the association of CENP-E with the microtubules. J. Biol. Chem. 2000;275:30451–30457. - PubMed
-
- Basto R., Scaerou F., Mische S., Wojcik E., Lefebvre C., Gomes R., Hays T., Karess R. In vivo dynamics of the rough deal checkpoint protein during Drosophila mitosis. Curr. Biol. 2004;14:56–61. - PubMed
-
- Busson S., Dujardin D., Moreau A., Dompierre J., De M., Jr Dynein and dynactin are localized to astral microtubules and at cortical sites in mitotic epithelial cells. Curr. Biol. 1998;8:541–544. - PubMed
-
- Chan G. K., Jablonski S. A., Starr D. A., Goldberg M. L., Yen T. J. Human Zw10 and ROD are mitotic checkpoint proteins that bind to kinetochores. Nat. Cell Biol. 2000;2:944–947. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

