Full genome characterisation of bluetongue virus serotype 6 from the Netherlands 2008 and comparison to other field and vaccine strains
- PMID: 20428242
- PMCID: PMC2859060
- DOI: 10.1371/journal.pone.0010323
Full genome characterisation of bluetongue virus serotype 6 from the Netherlands 2008 and comparison to other field and vaccine strains
Abstract
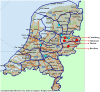
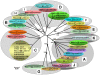
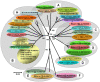
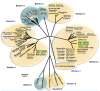
In mid September 2008, clinical signs of bluetongue (particularly coronitis) were observed in cows on three different farms in eastern Netherlands (Luttenberg, Heeten, and Barchem), two of which had been vaccinated with an inactivated BTV-8 vaccine (during May-June 2008). Bluetongue virus (BTV) infection was also detected on a fourth farm (Oldenzaal) in the same area while testing for export. BTV RNA was subsequently identified by real time RT-PCR targeting genome-segment (Seg-) 10, in blood samples from each farm. The virus was isolated from the Heeten sample (IAH "dsRNA virus reference collection" [dsRNA-VRC] isolate number NET2008/05) and typed as BTV-6 by RT-PCR targeting Seg-2. Sequencing confirmed the virus type, showing an identical Seg-2 sequence to that of the South African BTV-6 live-vaccine-strain. Although most of the other genome segments also showed very high levels of identity to the BTV-6 vaccine (99.7 to 100%), Seg-10 showed greatest identity (98.4%) to the BTV-2 vaccine (RSAvvv2/02), indicating that NET2008/05 had acquired a different Seg-10 by reassortment. Although Seg-7 from NET2008/05 was also most closely related to the BTV-6 vaccine (99.7/100% nt/aa identity), the Seg-7 sequence derived from the blood sample of the same animal (NET2008/06) was identical to that of the Netherlands BTV-8 (NET2006/04 and NET2007/01). This indicates that the blood contained two different Seg-7 sequences, one of which (from the BTV-6 vaccine) was selected during virus isolation in cell-culture. The predominance of the BTV-8 Seg-7 in the blood sample suggests that the virus was in the process of reassorting with the northern field strain of BTV-8. Two genome segments of the virus showed significant differences from the BTV-6 vaccine, indicating that they had been acquired by reassortment event with BTV-8, and another unknown parental-strain. However, the route by which BTV-6 and BTV-8 entered northern Europe was not established.
Conflict of interest statement
Figures


References
-
- Mertens PPC, Maan S, Samuel A, Attoui H. Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Orbivirus Reoviridae. . 2005. pp. 466–483. Virus Taxonomy VIIIth Report of the ICTV Elsevier/Academic Press London.
-
- Fernandez-Pacheco P, Fernandez-Pinero J, Aguero M, Jiminez-Clavero MA. Bluetongue virus serotype 1 in wild mouflons in Spain. Vet Rec. 2008;162:659–660. - PubMed
-
- Barzilai E, Tadmor A. Multiplication of bluetongue virus in goats following experimental infection. Refu Vet. 1971;28:11–20.
-
- Henrich M, Reinacher M, Hamann HP. Lethal bluetongue virus infection in an alpaca. Vet Rec. 2007;161:764. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

