Temporal characterization of homology-independent centromere coupling in meiotic prophase
- PMID: 20428251
- PMCID: PMC2859069
- DOI: 10.1371/journal.pone.0010336
Temporal characterization of homology-independent centromere coupling in meiotic prophase
Abstract
Background: Over the past thirty years several reports of the pairing or association of non-homologous centromeres during meiotic prophase have appeared in the literature. Recently, the homology-independent pairwise association of centromeres, termed centromere coupling, was also reported in budding yeast. It seems paradoxical that centromeres would pair with non-homologous partners during a process intended to align homologous chromosomes, yet the conservation of this phenomenon across a wide range of species suggests it may play an important role in meiosis.
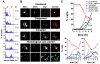
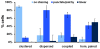
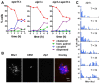
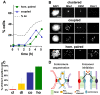
Principal findings: To better define the role of this phenomenon in budding yeast, experiments were preformed to place centromere coupling within the context of landmark meiotic events. Soon after the initiation of the meiotic program, centromeres were found to re-organize from a single cluster into non-homologous couples. Centromere coupling is detected as soon as chromosome replication is finished and persists while the recombination protein Dmc1 is loaded onto the chromosomes, suggesting that centromere coupling persists through the time of double strand break formation. In the absence of the synaptonemal complex component, Zip1, centromere coupling was undetectable, at all times examined, confirming the essential role of this protein on this process. Finally, the timely release of centromere coupling depends on the recombination-initiating enzyme, Spo11, suggesting a connection between events in homologous pairing/recombination and the regulation of centromere coupling.
Conclusions: Based on our results we propose a role for centromere coupling in blocking interactions between homologous centromeres as recombination initiation is taking place.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

