PoGO: Prediction of Gene Ontology terms for fungal proteins
- PMID: 20429880
- PMCID: PMC2882390
- DOI: 10.1186/1471-2105-11-215
PoGO: Prediction of Gene Ontology terms for fungal proteins
Abstract
Background: Automated protein function prediction methods are the only practical approach for assigning functions to genes obtained from model organisms. Many of the previously reported function annotation methods are of limited utility for fungal protein annotation. They are often trained only to one species, are not available for high-volume data processing, or require the use of data derived by experiments such as microarray analysis. To meet the increasing need for high throughput, automated annotation of fungal genomes, we have developed a tool for annotating fungal protein sequences with terms from the Gene Ontology.
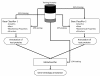
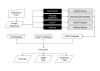
Results: We describe a classifier called PoGO (Prediction of Gene Ontology terms) that uses statistical pattern recognition methods to assign Gene Ontology (GO) terms to proteins from filamentous fungi. PoGO is organized as a meta-classifier in which each evidence source (sequence similarity, protein domains, protein structure and biochemical properties) is used to train independent base-level classifiers. The outputs of the base classifiers are used to train a meta-classifier, which provides the final assignment of GO terms. An independent classifier is trained for each GO term, making the system amenable to updating, without having to re-train the whole system. The resulting system is robust. It provides better accuracy and can assign GO terms to a higher percentage of unannotated protein sequences than other methods that we tested.
Conclusions: Our annotation system overcomes many of the shortcomings that we found in other methods. We also provide a web server where users can submit protein sequences to be annotated.
Figures
Similar articles
-
Gene Ontology annotation of the rice blast fungus, Magnaporthe oryzae.BMC Microbiol. 2009 Feb 19;9 Suppl 1(Suppl 1):S8. doi: 10.1186/1471-2180-9-S1-S8. BMC Microbiol. 2009. PMID: 19278556 Free PMC article. Review.
-
ProLoc-GO: utilizing informative Gene Ontology terms for sequence-based prediction of protein subcellular localization.BMC Bioinformatics. 2008 Feb 1;9:80. doi: 10.1186/1471-2105-9-80. BMC Bioinformatics. 2008. PMID: 18241343 Free PMC article.
-
Protein function prediction using text-based features extracted from the biomedical literature: the CAFA challenge.BMC Bioinformatics. 2013;14 Suppl 3(Suppl 3):S14. doi: 10.1186/1471-2105-14-S3-S14. Epub 2013 Feb 28. BMC Bioinformatics. 2013. PMID: 23514326 Free PMC article.
-
MIPS: analysis and annotation of genome information in 2007.Nucleic Acids Res. 2008 Jan;36(Database issue):D196-201. doi: 10.1093/nar/gkm980. Epub 2007 Dec 23. Nucleic Acids Res. 2008. PMID: 18158298 Free PMC article.
-
Comparing genomes in terms of protein structure: surveys of a finite parts list.FEMS Microbiol Rev. 1998 Oct;22(4):277-304. doi: 10.1111/j.1574-6976.1998.tb00371.x. FEMS Microbiol Rev. 1998. PMID: 10357579 Review.
Cited by
-
Protein function prediction with gene ontology: from traditional to deep learning models.PeerJ. 2021 Aug 24;9:e12019. doi: 10.7717/peerj.12019. eCollection 2021. PeerJ. 2021. PMID: 34513334 Free PMC article.
-
Predictability of gene ontology slim-terms from primary structure information in Embryophyta plant proteins.BMC Bioinformatics. 2013 Feb 26;14:68. doi: 10.1186/1471-2105-14-68. BMC Bioinformatics. 2013. PMID: 23441934 Free PMC article.
-
Review on the Computational Genome Annotation of Sequences Obtained by Next-Generation Sequencing.Biology (Basel). 2020 Sep 18;9(9):295. doi: 10.3390/biology9090295. Biology (Basel). 2020. PMID: 32962098 Free PMC article. Review.
-
Genome-wide identification and comprehensive analyses of the kinomes in four pathogenic microsporidia species.PLoS One. 2014 Dec 30;9(12):e115890. doi: 10.1371/journal.pone.0115890. eCollection 2014. PLoS One. 2014. PMID: 25549259 Free PMC article.
-
CvManGO, a method for leveraging computational predictions to improve literature-based Gene Ontology annotations.Database (Oxford). 2012 Mar 20;2012:bas001. doi: 10.1093/database/bas001. Print 2012. Database (Oxford). 2012. PMID: 22434836 Free PMC article.
References
-
- King RD, Karwath A, Clare A, Dephaspe L. Genome scale prediction of protein functional class from sequence using data mining. Proc of the sixth ACM SIGKDD Inter Conf on Knowledge discovery and data mining. 2003.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

