Bayesian shrinkage mapping of quantitative trait loci in variance component models
- PMID: 20429900
- PMCID: PMC2874758
- DOI: 10.1186/1471-2156-11-30
Bayesian shrinkage mapping of quantitative trait loci in variance component models
Abstract
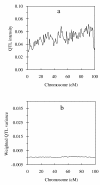
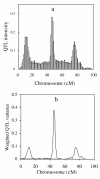
Background: In this article, I propose a model-selection-free method to map multiple quantitative trait loci (QTL) in variance component model, which is useful in outbred populations. The new method can estimate the variance of zero-effect QTL infinitely to zero, but nearly unbiased for non-zero-effect QTL. It is analogous to Xu's Bayesian shrinkage estimation method, but his method is based on allelic substitution model, while the new method is based on the variance component models.
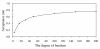
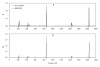
Results: Extensive simulation experiments were conducted to investigate the performance of the proposed method. The results showed that the proposed method was efficient in mapping multiple QTL simultaneously, and moreover it was more competitive than the reversible jump MCMC (RJMCMC) method and may even out-perform it.
Conclusions: The newly developed Bayesian shrinkage method is very efficient and powerful for mapping multiple QTL in outbred populations.
Figures


Similar articles
-
A new Bayesian automatic model selection approach for mapping quantitative trait loci under variance component model.Genetica. 2009 Apr;135(3):429-37. doi: 10.1007/s10709-008-9291-5. Epub 2008 Jul 22. Genetica. 2009. PMID: 18648990
-
QTL mapping in outbred half-sib families using Bayesian model selection.Heredity (Edinb). 2011 Sep;107(3):265-76. doi: 10.1038/hdy.2011.15. Epub 2011 Apr 13. Heredity (Edinb). 2011. PMID: 21487433 Free PMC article.
-
Bayesian mapping of quantitative trait loci for multiple complex traits with the use of variance components.Am J Hum Genet. 2007 Aug;81(2):304-20. doi: 10.1086/519495. Epub 2007 Jul 3. Am J Hum Genet. 2007. PMID: 17668380 Free PMC article.
-
Overview of LASSO-related penalized regression methods for quantitative trait mapping and genomic selection.Theor Appl Genet. 2012 Aug;125(3):419-35. doi: 10.1007/s00122-012-1892-9. Epub 2012 May 24. Theor Appl Genet. 2012. PMID: 22622521 Review.
-
Advances in Bayesian multiple quantitative trait loci mapping in experimental crosses.Heredity (Edinb). 2008 Mar;100(3):240-52. doi: 10.1038/sj.hdy.6801074. Epub 2007 Nov 7. Heredity (Edinb). 2008. PMID: 17987056 Free PMC article. Review.
Cited by
-
Association analysis for udder health based on SNP-panel and sequence data in Danish Holsteins.Genet Sel Evol. 2015 Jun 19;47(1):50. doi: 10.1186/s12711-015-0129-1. Genet Sel Evol. 2015. PMID: 26087655 Free PMC article.
-
A new mapping method for quantitative trait loci of silkworm.BMC Genet. 2011 Jan 28;12:19. doi: 10.1186/1471-2156-12-19. BMC Genet. 2011. PMID: 21276233 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

