CLOCK regulates circadian rhythms of hepatic glycogen synthesis through transcriptional activation of Gys2
- PMID: 20430893
- PMCID: PMC2903367
- DOI: 10.1074/jbc.M110.110361
CLOCK regulates circadian rhythms of hepatic glycogen synthesis through transcriptional activation of Gys2
Abstract
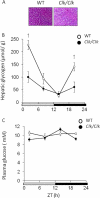
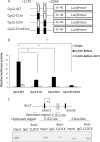
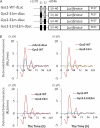
Hepatic glycogen content is important for glucose homeostasis and exhibits robust circadian rhythms that peak at the end of the active phase in mammals. The activities of the rate-limiting enzymes for glycogenesis and glycogenolysis also show circadian rhythms, and the balance between them forms the circadian rhythm of the hepatic glycogen content. However, no direct evidence has yet implicated the circadian clock in the regulation of glycogen metabolism at the molecular level. We show here that a Clock gene mutation damps the circadian rhythm of the hepatic glycogen content, as well as the circadian mRNA and protein expression of Gys2 (glycogen synthase 2), which is the rate-limiting enzyme of glycogenesis in the liver. Transient reporter assays revealed that CLOCK drives the transcriptional activation of Gys2 via two tandemly located E-boxes. Chromatin immunoprecipitation assays of liver tissues revealed that CLOCK binds to these E-box elements in vivo, and real time reporter assays showed that these elements are sufficient for circadian Gys2 expression in vitro. Thus, CLOCK regulates the circadian rhythms of hepatic glycogen synthesis through transcriptional activation of Gys2.
Figures



References
-
- Pando M. P., Sassone-Corsi P. (2001) Nature 410, 311–313 - PubMed
-
- Reppert S. M., Weaver D. R. (2001) Annu. Rev. Physiol. 63, 647–676 - PubMed
-
- Sakamoto K., Nagase T., Fukui H., Horikawa K., Okada T., Tanaka H., Sato K., Miyake Y., Ohara O., Kako K., Ishida N. (1998) J. Biol. Chem. 273, 27039–27042 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

