Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations: diagnostic and clinical implications
- PMID: 20431034
- PMCID: PMC2893626
- DOI: 10.2353/jmoldx.2010.090188
Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations: diagnostic and clinical implications
Abstract
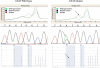
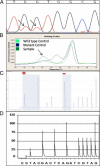
Mutations in codons 12 and 13 of the KRAS oncogene are relatively common in colorectal and lung adenocarcinomas. Recent data indicate that these mutations result in resistance to anti-epidermal growth factor receptor therapy. Therefore, we assessed Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS codon 12/13 mutations in formalin-fixed paraffin-embedded samples, including 58 primary and 42 metastatic colorectal adenocarcinomas, 63 primary and 17 metastatic lung adenocarcinomas, and 20 normal colon samples. Of 180 tumor samples, 62.2% were KRAS mutant positive, and 37.8% were negative. Melting curve analysis yielded no false positive or false negative results, but had 10% equivocal calls. Melting curve analysis also resulted in 4 cases with melting curves inconsistent with either wild-type or codon 12/13 mutations. These patterns were generated from samples with double mutants in codons 12/13 and with mutations outside of codons 12/13. Pyrosequencing yielded no false positive or false negative results as well. However, two samples from one patient yielded a pyrogram that was flagged as abnormal, but the mutation subtype could not be determined. Finally, using an electronic cutoff of 10%, Sanger sequencing showed 11.1% false positives and 6.1% false negatives. In our hands, the limit of detection for Sanger sequencing, pyrosequencing, and melting curve analysis was approximately 15 to 20%, 5%, and 10% mutant alleles, respectively.
Figures

References
-
- Ellis RW, Defeo D, Shih TY, Gonda MA, Young HA, Tsuchida N, Lowy DR, Scolnick EM. The p21 src genes of Harvey and Kirsten sarcoma viruses originate from divergent members of a family of normal vertebrate genes. Nature. 1981;292:506–511. - PubMed
-
- Bos JL, Fearon ER, Hamilton SR, Verlaan-de Vries M, van Boom JH, van der Eb AJ, Vogelstein B. Prevalence of ras gene mutations in human colorectal cancers. Nature. 1987;327:293–297. - PubMed
-
- Minamoto T, Mai M, Ronai Z. K-ras mutation: early detection in molecular diagnosis and risk assessment of colorectal, pancreas, and lung cancers–a review. Cancer Detect Prev. 2000;24:1–12. - PubMed
-
- Der CJ, Cooper GM. Altered gene products are associated with activation of cellular rasK genes in human lung and colon carcinomas. Cell. 1983;32:201–208. - PubMed
-
- Sugio K, Kishimoto Y, Virmani AK, Hung JY, Gazdar AF. K-ras mutations are a relatively late event in the pathogenesis of lung carcinomas. Cancer Res. 1994;54:5811–5815. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

