Studies of a biochemical factory: tomato trichome deep expressed sequence tag sequencing and proteomics
- PMID: 20431087
- PMCID: PMC2899918
- DOI: 10.1104/pp.110.157214
Studies of a biochemical factory: tomato trichome deep expressed sequence tag sequencing and proteomics
Abstract
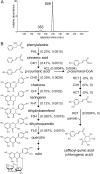
Shotgun proteomics analysis allows hundreds of proteins to be identified and quantified from a single sample at relatively low cost. Extensive DNA sequence information is a prerequisite for shotgun proteomics, and it is ideal to have sequence for the organism being studied rather than from related species or accessions. While this requirement has limited the set of organisms that are candidates for this approach, next generation sequencing technologies make it feasible to obtain deep DNA sequence coverage from any organism. As part of our studies of specialized (secondary) metabolism in tomato (Solanum lycopersicum) trichomes, 454 sequencing of cDNA was combined with shotgun proteomics analyses to obtain in-depth profiles of genes and proteins expressed in leaf and stem glandular trichomes of 3-week-old plants. The expressed sequence tag and proteomics data sets combined with metabolite analysis led to the discovery and characterization of a sesquiterpene synthase that produces beta-caryophyllene and alpha-humulene from E,E-farnesyl diphosphate in trichomes of leaf but not of stem. This analysis demonstrates the utility of combining high-throughput cDNA sequencing with proteomics experiments in a target tissue. These data can be used for dissection of other biochemical processes in these specialized epidermal cells.
Figures



Similar articles
-
Role of trichomes in defense against herbivores: comparison of herbivore response to woolly and hairless trichome mutants in tomato (Solanum lycopersicum).Planta. 2012 Oct;236(4):1053-66. doi: 10.1007/s00425-012-1651-9. Epub 2012 May 3. Planta. 2012. PMID: 22552638
-
Engineering of Tomato Glandular Trichomes for the Production of Specialized Metabolites.Methods Enzymol. 2016;576:305-31. doi: 10.1016/bs.mie.2016.02.014. Epub 2016 Mar 24. Methods Enzymol. 2016. PMID: 27480691
-
RNA-seq discovery, functional characterization, and comparison of sesquiterpene synthases from Solanum lycopersicum and Solanum habrochaites trichomes.Plant Mol Biol. 2011 Nov;77(4-5):323-36. doi: 10.1007/s11103-011-9813-x. Epub 2011 Aug 5. Plant Mol Biol. 2011. PMID: 21818683 Free PMC article.
-
Glandular trichomes: micro-organs with model status?New Phytol. 2020 Mar;225(6):2251-2266. doi: 10.1111/nph.16283. Epub 2019 Dec 10. New Phytol. 2020. PMID: 31651036 Review.
-
Glandular trichomes: what comes after expressed sequence tags?Plant J. 2012 Apr;70(1):51-68. doi: 10.1111/j.1365-313X.2012.04913.x. Plant J. 2012. PMID: 22449043 Review.
Cited by
-
Cannabis sativa: The Plant of the Thousand and One Molecules.Front Plant Sci. 2016 Feb 4;7:19. doi: 10.3389/fpls.2016.00019. eCollection 2016. Front Plant Sci. 2016. PMID: 26870049 Free PMC article. Review.
-
Acylsugar Acylhydrolases: Carboxylesterase-Catalyzed Hydrolysis of Acylsugars in Tomato Trichomes.Plant Physiol. 2016 Mar;170(3):1331-44. doi: 10.1104/pp.15.01348. Epub 2016 Jan 25. Plant Physiol. 2016. PMID: 26811191 Free PMC article.
-
Using Gene Expression to Study Specialized Metabolism-A Practical Guide.Front Plant Sci. 2021 Jan 12;11:625035. doi: 10.3389/fpls.2020.625035. eCollection 2020. Front Plant Sci. 2021. PMID: 33510763 Free PMC article. Review.
-
The SlGRAS9-SlMYC1 regulatory module controls glandular trichome formation and modulates resilience to pest in tomato.Plant J. 2025 May;122(3):e70183. doi: 10.1111/tpj.70183. Plant J. 2025. PMID: 40344466 Free PMC article.
-
Mono- and sesquiterpene release from tomato (Solanum lycopersicum) leaves upon mild and severe heat stress and through recovery: from gene expression to emission responses.Environ Exp Bot. 2016 Dec;132:1-15. doi: 10.1016/j.envexpbot.2016.08.003. Environ Exp Bot. 2016. PMID: 29367791 Free PMC article.
References
-
- Alvarez S, Marsh EL, Schroeder SG, Schachtman DP. (2008) Metabolomic and proteomic changes in the xylem sap of maize under drought. Plant Cell Environ 31: 325–340 - PubMed
-
- Amme S, Rutten T, Melzer M, Sonsmann G, Vissers JPC, Schlesier B, Mock HP. (2005) A proteome approach defines protective functions of tobacco leaf trichomes. Proteomics 5: 2508–2518 - PubMed
-
- Bertea CM, Voster A, Verstappen FWA, Maffei M, Beekwilder J, Bouwmeester HJ. (2006) Isoprenoid biosynthesis in Artemisia annua: cloning and heterologous expression of a germacrene A synthase from a glandular trichome cDNA library. Arch Biochem Biophys 448: 3–12 - PubMed
-
- Bräutigam A, Shrestha RP, Whitten D, Wilkerson CG, Carr KM, Froehlich JE, Weber APM. (2008) Low-coverage massively parallel pyrosequencing of cDNAs enables proteomics in non-model species: comparison of a species-specific database generated by pyrosequencing with databases from related species for proteome analysis of pea chloroplast envelopes. J Biotechnol 136: 44–53 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

