The Groucho ortholog UNC-37 interacts with the short Groucho-like protein LSY-22 to control developmental decisions in C. elegans
- PMID: 20431118
- PMCID: PMC2867316
- DOI: 10.1242/dev.046219
The Groucho ortholog UNC-37 interacts with the short Groucho-like protein LSY-22 to control developmental decisions in C. elegans
Abstract
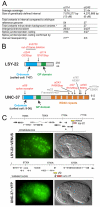
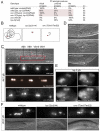
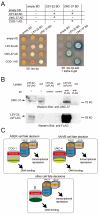
Transcriptional co-repressors of the Groucho/TLE family are important regulators of development in many species. A subset of Groucho/TLE family members that lack the C-terminal WD40 domains have been proposed to act as dominant-negative regulators of Groucho/TLE proteins, yet such a role has not been conclusively proven. Through a mutant screen for genes controlling a left/right asymmetric cell fate decision in the nervous system of the nematode C. elegans, we have retrieved loss-of-function alleles in two distinct loci that display identical phenotypes in neuronal fate specification and in other developmental contexts. Using the novel technology of whole-genome sequencing, we find that these loci encode the C. elegans ortholog of Groucho, UNC-37, and, surprisingly, a short Groucho-like protein, LSY-22, that is similar to truncated Groucho proteins in other species. Besides their phenotypic similarities, unc-37 and lsy-22 show genetic interactions and UNC-37 and LSY-22 proteins also physically bind to each other in vivo. Our findings suggest that rather than acting as negative regulators of Groucho, small Groucho-like proteins may promote Groucho function. We propose that Groucho-mediated gene regulatory events involve heteromeric complexes of distinct Groucho-like proteins.
Figures

References
-
- Blelloch R., Anna-Arriola S. S., Gao D., Li Y., Hodgkin J., Kimble J. (1999). The gon-1 gene is required for gonadal morphogenesis in Caenorhabditis elegans. Dev. Biol. 216, 382-393 - PubMed
-
- Buscarlet M., Stifani S. (2007). The `Marx' of Groucho on development and disease. Trends Cell Biol. 17, 353-361 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

