Histone chaperones, histone acetylation, and the fluidity of the chromogenome
- PMID: 20432449
- PMCID: PMC3184832
- DOI: 10.1002/jcp.22150
Histone chaperones, histone acetylation, and the fluidity of the chromogenome
Abstract
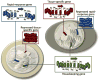
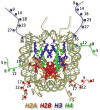
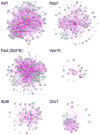
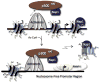
The "chromogenome" is defined as the structural and functional status of the genome at any given moment within a eukaryotic cell. This article focuses on recently uncovered relationships between histone chaperones, post-translational acetylation of histones, and modulation of the chromogenome. We emphasize those chaperones that function in a replication-independent manner, and for which three-dimensional structural information has been obtained. The emerging links between histone acetylation and chaperone function in both yeast and higher metazoans are discussed, including the importance of nucleosome-free regions. We close by posing many questions pertaining to how the coupled action of histone chaperones and acetylation influences chromogenome structure and function.
Figures


References
-
- Adkins MW, Howar SR, Tyler JK. Chromatin Disassembly Mediated by the Histone Chaperone Asf1 Is Essential for Transcriptional Activation of the Yeast PHO5 and PHO8 Genes. Mol Cell. 2004;14(5):657–666. - PubMed
-
- Adkins MW, Tyler JK. Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Mol Cell. 2006;21(3):405–416. - PubMed
-
- Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication- independent nucleosome assembly. Mol Cell. 2002;9(6):1191–1200. - PubMed
-
- Akey CW, Luger K. Histone chaperones and nucleosome assembly. Curr Opin Struct Biol. 2003;13(1):6–14. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

