Rapid targeted mutational analysis of human tumours: a clinical platform to guide personalized cancer medicine
- PMID: 20432502
- PMCID: PMC3377316
- DOI: 10.1002/emmm.201000070
Rapid targeted mutational analysis of human tumours: a clinical platform to guide personalized cancer medicine
Abstract
Targeted cancer therapy requires the rapid and accurate identification of genetic abnormalities predictive of therapeutic response. We sought to develop a high-throughput genotyping platform that would allow prospective patient selection to the best available therapies, and that could readily and inexpensively be adopted by most clinical laboratories. We developed a highly sensitive multiplexed clinical assay that performs very well with nucleic acid derived from formalin fixation and paraffin embedding (FFPE) tissue, and tests for 120 previously described mutations in 13 cancer genes. Genetic profiling of 250 primary tumours was consistent with the documented oncogene mutational spectrum and identified rare events in some cancer types. The assay is currently being used for clinical testing of tumour samples and contributing to cancer patient management. This work therefore establishes a platform for real-time targeted genotyping that can be widely adopted. We expect that efforts like this one will play an increasingly important role in cancer management.
Figures

The SNaPshot system follows a straightforward protocol and uses infrastructure already existent in most clinical laboratories. This method consists of a multiplexed PCR step, followed by a single-base extension sequencing reaction, in which allele-specific probes interrogate loci of interest and are fluorescently labelled using dideoxynucleotides. These probes are designed to have different sizes and are subsequently resolved by electrophoresis and analysed by an automated DNA sequencer. Thus, the identity of each locus is given by the position of its corresponding fluorescent peak in the spectrum, which is dictated by the length of the extension primer.
Detailed view of the single-base extension reaction. The identity of the nucleotide(s) present at each locus is given by two parameters: the molecular weight and the colour of the fluorescently labelled ddNTPs added to the allele specific probes during the extension step. Thus, mutant and wild-type alleles can be distinguished based on the slightly different positions and on the distinct colours of their corresponding peaks. These two factors are used to establish the bins used for automatic data analysis (described in the Supporting Information).

The A427 lung carcinoma cell line was used to detect the KRAS G12D mutation (nucleotide change 35G > A). Sensitivity was ∼3% and the SNaPshot panel includes the following assays: (1) KRAS 35; (2) EGFR 2236_50del R; (3) PTEN 517; (4) TP53 733; (5) FLT3 2503; (6) PIK3CA 3139; (7) NOTCH1 4724 and (8) NOTCH1 4802.
The NCI-H1975 lung adenocarcinoma cell line was used to identify the EGFR T790M mutation (nucleotide change 2369C > T). Assay sensitivity was ∼3% and the SNaPshot panel tests for: (1) KRAS 34; (2) EGFR 2235_49del F; (3) EGFR 2369; (4) NRAS 181; (5) PIK3CA 1633; (6) CTNNB1 94 and (7) CTNNB1 121. As can be appreciated in the middle section, decreasing levels of ‘green’ mutant signal (arrows), absent from wild-type DNA (top panel), can be easily distinguished from the nearby ‘red’ background peak (asterisk), which is also found in the assay run on the normal control (top panel). Of note, the EGFR c.2369C assay was designed in the reverse orientation, thus the observed alleles are G (blue) for the wild-type and A (green) for the mutant.
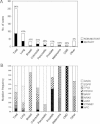
their mutational status and
the mutation frequency of individual genes.
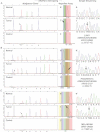
Detection of the EGFR L858R (c.2573T > G) mutation in a case of lung adenocarcinoma. Assays: (1) EGFR 2236_50del F; (2) EGFR 2573; (3) CTNNB1 133; (4) PIK3CA 1624 and (5) NRAS 35.
Identification of the KRAS G12V (c.35G > T) mutation in a pancreatic adenocarcinoma. Assays: (1) KRAS 35; (2) EGFR 2236_50del R; (3) PTEN 517; (4) TP53 733; (5) FLT3 2503; (6) PIK3CA 3139; (7) NOTCH1 4724 and (8) NOTCH1 4802.
Detection of the BRAF V600E (c.1799T > A) mutation in melanoma. Assays: (1) EGFR 2235_49del R; (2) NRAS 38; (3) BRAF 1799; (4) NRAS 182; (5) PIK3CA 263; (6) TP53 742; (7) CTNNB1 95 and (8) CTNNB1 122.
Comment in
-
Mutations for the people.EMBO Mol Med. 2010 May;2(5):143-5. doi: 10.1002/emmm.201000071. EMBO Mol Med. 2010. PMID: 20461736 Free PMC article.
References
-
- Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K, Linn SC, Gonzalez-Angulo AM, Stemke-Hale K, Hauptmann M, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. - PubMed
-
- Davies H, Hunter C, Smith R, Stephens P, Greenman C, Bignell G, Teague J, Butler A, Edkins S, Stevens C, et al. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res. 2005;65:7591–7595. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

