Genomic instability in breast cancer: pathogenesis and clinical implications
- PMID: 20434415
- PMCID: PMC2904860
- DOI: 10.1016/j.molonc.2010.04.001
Genomic instability in breast cancer: pathogenesis and clinical implications
Abstract
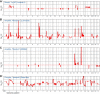
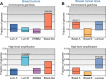
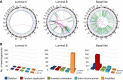
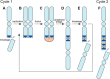
Breast cancer is a heterogeneous disease, appreciable by molecular markers, gene-expression profiles, and most recently, patterns of genomic alteration. In particular, genomic profiling has revealed three distinct patterns of DNA copy-number alteration: a "simple" type with few gains or losses of whole chromosome arms, an "amplifier" type with focal high-level DNA amplifications, and a "complex" type marked by numerous low-amplitude changes and copy-number transitions. The three patterns are associated with distinct gene-expression subtypes, and preferentially target different loci in the genome (implicating distinct cancer genes). Moreover, the different patterns of alteration imply distinct underlying mechanisms of genomic instability. The amplifier pattern may arise from transient telomere dysfunction, although new data suggest ongoing "amplifier" instability. The complex pattern shows similarity to breast cancers with germline BRCA1 mutation, which also exhibit "basal-like" expression profiles and complex-pattern genomes, implicating a possible defect in BRCA1-associated repair of DNA double-strand breaks. As such, targeting presumptive DNA repair defects represents a promising area of clinical investigation. Future studies should clarify the pathogenesis of breast cancers with amplifier and complex-pattern genomes, and will likely identify new therapeutic opportunities.
(c) 2010 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Figures
References
-
- Albertson, D.G. , 2006. Gene amplification in cancer. Trends Genet. 22, 447–455. - PubMed
-
- Artandi, S.E. , DePinho, R.A. , 2000. A critical role for telomeres in suppressing and facilitating carcinogenesis. Curr. Opin. Genet. Dev. 10, 39–46. - PubMed
-
- Bergamaschi, A. , Kim, Y.H. , Wang, P. , Sorlie, T. , Hernandez-Boussard, T. , Lonning, P.E. , Tibshirani, R. , Borresen-Dale, A.L. , Pollack, J.R. , 2006. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 45, 1033–1040. - PubMed
-
- van Beers, E.H. , van Welsem, T. , Wessels, L.F. , Li, Y. , Oldenburg, R.A. , Devilee, P. , Cornelisse, C.J. , Verhoef, S. , Hogervorst, F.B. , van't Veer, L.J. , Nederlof, P.M. , 2005. Comparative genomic hybridization profiles in human BRCA1 and BRCA2 breast tumors highlight differential sets of genomic aberrations. Cancer Res. 65, 822–827. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

