Titration-free massively parallel pyrosequencing using trace amounts of starting material
- PMID: 20435675
- PMCID: PMC2910068
- DOI: 10.1093/nar/gkq332
Titration-free massively parallel pyrosequencing using trace amounts of starting material
Abstract
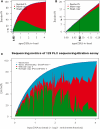
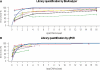
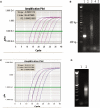
Continuous efforts have been made to improve next-generation sequencing methods for increased robustness and for applications on low amounts of starting material. We applied double-stranded library protocols for the Roche 454 platform to avoid the yield-reducing steps associated with single-stranded library preparation, and applied a highly sensitive Taqman MGB-probe-based quantitative polymerase chain reaction (qPCR) method. The MGB-probe qPCR, which can detect as low as 100 copies, was used to quantify the amount of effective library, i.e. molecules that form functional clones in emulsion PCR. We also demonstrate that the distribution of library molecules on capture beads follows a Poisson distribution. Combining the qPCR and Poisson statistics, the labour-intensive and costly titration can be eliminated and trace amounts of starting material such as precious clinical samples, transcriptomes of small tissue samples and metagenomics on low biomass environments is applicable.
Figures
References
-
- Harris TD, Buzby PR, Babcock H, Beer E, Bowers J, Braslavsky I, Causey M, Colonell J, Dimeo J, Efcavitch JW, et al. Single-molecule DNA sequencing of a viral genome. Science. 2008;320:106–109. - PubMed
-
- Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. - PubMed
-
- Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S, Bayley H. Continuous base identification for single-molecule nanopore DNA sequencing. Nat. Nanotechnol. 2009;4:265–270. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

