ConPlex: a server for the evolutionary conservation analysis of protein complex structures
- PMID: 20435678
- PMCID: PMC2896159
- DOI: 10.1093/nar/gkq328
ConPlex: a server for the evolutionary conservation analysis of protein complex structures
Abstract
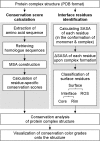
Evolutionary conservation analyses are important for the identification of protein-protein interactions. For protein complex structures, sequence conservation has been applied to determine protein oligomerization states, to characterize native interfaces from non-specific crystal contacts, and to discriminate near-native structures from docking artifacts. However, a user-friendly web-based service for evolutionary conservation analysis of protein complexes has not been available. Therefore, we developed ConPlex (http://sbi.postech.ac.kr/ConPlex/) a web application that enables evolutionary conservation analyses of protein interactions within protein quaternary structures. Users provide protein complex structures; ConPlex automatically identifies protein interfaces and carries out evolutionary conservation analyses for the interface regions. Moreover, ConPlex allows the results of the residue-specific conservation analysis to be displayed on the protein complex structure and provides several options to customize the display output to fit each user's needs. We believe that ConPlex offers a convenient platform to analyze protein complex structures based on evolutionary conservation of protein-protein interface residues.
Figures


References
-
- Pupko T, Bell RE, Mayrose I, Glaser F, Ben-Tal N. Rate4Site: an algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics. 2002;18(Suppl. 1):S71–S77. - PubMed
-
- Bradford JR, Westhead DR. Improved prediction of protein-protein binding sites using a support vector machines approach. Bioinformatics. 2005;21:1487–1494. - PubMed
-
- Berezin C, Glaser F, Rosenberg J, Paz I, Pupko T, Fariselli P, Casadio R, Ben-Tal N. ConSeq: the identification of functionally and structurally important residues in protein sequences. Bioinformatics. 2004;20:1322–1324. - PubMed
-
- Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben-Tal N. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

