Characterization of the branched-chain amino acid aminotransferase enzyme family in tomato
- PMID: 20435740
- PMCID: PMC2899903
- DOI: 10.1104/pp.110.154922
Characterization of the branched-chain amino acid aminotransferase enzyme family in tomato
Abstract
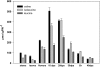
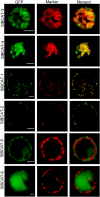
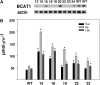
Branched-chain amino acids (BCAAs) are synthesized in plants from branched-chain keto acids, but their metabolism is not completely understood. The interface of BCAA metabolism lies with branched-chain aminotransferases (BCAT) that catalyze both the last anabolic step and the first catabolic step. In this study, six BCAT genes from the cultivated tomato (Solanum lycopersicum) were identified and characterized. SlBCAT1, -2, -3, and -4 are expressed in multiple plant tissues, while SlBCAT5 and -6 were undetectable. SlBCAT1 and -2 are located in the mitochondria, SlBCAT3 and -4 are located in chloroplasts, while SlBCAT5 and -6 are located in the cytosol and vacuole, respectively. SlBCAT1, -2, -3, and -4 were able to restore growth of Escherichia coli BCAA auxotrophic cells, but SlBCAT1 and -2 were less effective than SlBCAT3 and -4 in growth restoration. All enzymes were active in the forward (BCAA synthesis) and reverse (branched-chain keto acid synthesis) reactions. SlBCAT3 and -4 exhibited a preference for the forward reaction, while SlBCAT1 and -2 were more active in the reverse reaction. While overexpression of SlBCAT1 or -3 in tomato fruit did not significantly alter amino acid levels, an expression quantitative trait locus on chromosome 3, associated with substantially higher expression of Solanum pennellii BCAT4, did significantly increase BCAA levels. Conversely, antisense-mediated reduction of SlBCAT1 resulted in higher levels of BCAAs. Together, these results support a model in which the mitochondrial SlBCAT1 and -2 function in BCAA catabolism while the chloroplastic SlBCAT3 and -4 function in BCAA synthesis.
Figures


References
-
- Beck HC, Hansen AM, Lauritsen FR. (2004) Catabolism of leucine to branched-chain fatty acids in Staphylococcus xylosus. J Appl Microbiol 96: 1185–1193 - PubMed
-
- Binder S, Knill T, Schuster J. (2007) Branched-chain amino acid metabolism in higher plants. Physiol Plant 129: 68–78
-
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 - PubMed
-
- Carrari F, Baxter C, Usadel B, Urbanczyk-Wochniak E, Zanor MI, Nunes-Nesi A, Nikiforova V, Centeno DC, Ratzka A, Pauly M, et al. (2006) Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiol 142: 1380–1396 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

