Commonalities and differences in regulation of N-acyl homoserine lactone quorum sensing in the beneficial plant-associated burkholderia species cluster
- PMID: 20435760
- PMCID: PMC2897448
- DOI: 10.1128/AEM.03086-09
Commonalities and differences in regulation of N-acyl homoserine lactone quorum sensing in the beneficial plant-associated burkholderia species cluster
Abstract
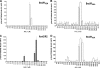
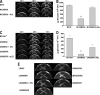
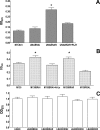
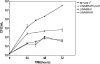
The genus Burkholderia includes over 60 species isolated from a wide range of environmental niches and can be tentatively divided into two major species clusters. The first cluster includes pathogens such as Burkholderia glumae, B. pseudomallei, and B. mallei and 17 well-studied species of the Burkholderia cepacia complex. The other recently established cluster comprises at least 29 nonpathogenic species, which in most cases have been found to be associated with plants. It was previously established that Burkholderia kururiensis, a member of the latter cluster, possesses an N-acyl homoserine lactone (AHL) quorum-sensing (QS) system designated "BraI/R," which is found in all species of the plant-associated cluster. In the present study, two other BraI/R-like systems were characterized in B. xenovorans and B. unamae and were designated the BraI/R(XEN) and BraI/R(UNA) systems, respectively. Several phenotypes were analyzed, and it was determined that exopolysaccharide was positively regulated by the BraIR-like system in the species B. kururiensis, B. unamae, and B. xenovorans, highlighting commonality in targets. However, the three BraIR-like systems also revealed differences in targets since biofilm formation and plant colonization were differentially regulated. In addition, a second AHL QS system designated XenI2/R2 and an unpaired LuxR solo protein designated BxeR solo were also identified and characterized in B. xenovorans LB400(T). The two AHL QS systems of B. xenovorans are not transcriptionally regulating each other, whereas BxeR solo negatively regulated xenI2. The XenI2/R2 and BxeR solo proteins are not widespread in the Burkholderia species cluster. In conclusion, the present study represents an extensive analysis of AHL QS in the Burkholderia plant-associated cluster demonstrating both commonalities and differences, probably reflecting environmental adaptations of the various species.
Figures



References
-
- Alexeyev, M. F. 1999. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. Biotechniques 26:824-828. - PubMed
-
- Anderson, J. A. 1939. The use of tributyrin agar in dairy bacteriology. Berlin Int. Mikrobiol. Kongress 3:726-728.
-
- Arevalo-Ferro, C., M. Hentzer, G. Reil, A. Gorg, S. Kjelleberg, M. Givskov, K. Riedel, and L. Eberl. 2003. Identification of quorum-sensing regulated proteins in the opportunistic pathogen Pseudomonas aeruginosa by proteomics. Environ. Microbiol. 5:1350-1369. - PubMed
-
- Baldani, V. L., E. Oliveira, E. Balota, J. I. Baldani, G. Kirchhof, and J. Döbereiner. 1997. Burkholderia brasilensis sp. nov. uma nova espécie de bactéria diazotrófica endofítica. An. Acad. Bras. Cienc. 69:1.
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

