Direct interactions of intraflagellar transport complex B proteins IFT88, IFT52, and IFT46
- PMID: 20435895
- PMCID: PMC2898376
- DOI: 10.1074/jbc.M110.106997
Direct interactions of intraflagellar transport complex B proteins IFT88, IFT52, and IFT46
Abstract
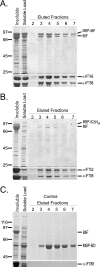
Intraflagellar transport (IFT) particles of Chlamydomonas reinhardtii contain two distinct protein complexes, A and B, composed of at least 6 and 15 protein subunits, respectively. As isolated from C. reinhardtii flagella, IFT complex B can be further reduced to a approximately 500-kDa core that contains IFT88, 2x IFT81, 2x IFT74/72, IFT52, IFT46, IFT27, IFT25, and IFT22. In this study, yeast-based two-hybrid analysis was combined with bacterial coexpression to show that three of the core B subunits, IFT88, IFT52, and IFT46, interact directly with each other and, together, are capable of forming a ternary complex. Chemical cross-linking results support the IFT52-IFT88 interaction and provide additional evidence of an association between IFT27 and IFT81. With previous studies showing that IFT81 and IFT74/72 interact to form a (IFT81)(2)(IFT74/72)(2) heterotetramer and that IFT27 and IFT25 form a heterodimer, the architecture of complex B is revealing itself. Last, electroporation of recombinant IFT46 was used to rescue flagellar assembly of a newly identified ift46 mutant and to monitor in vivo localization and movement of the IFT particles.
Figures





Similar articles
-
Characterization of the intraflagellar transport complex B core: direct interaction of the IFT81 and IFT74/72 subunits.J Biol Chem. 2005 Jul 29;280(30):27688-96. doi: 10.1074/jbc.M505062200. Epub 2005 Jun 13. J Biol Chem. 2005. PMID: 15955805
-
Intraflagellar transport protein IFT52 recruits IFT46 to the basal body and flagella.J Cell Sci. 2017 May 1;130(9):1662-1674. doi: 10.1242/jcs.200758. Epub 2017 Mar 16. J Cell Sci. 2017. PMID: 28302912
-
Dissecting the sequential assembly and localization of intraflagellar transport particle complex B in Chlamydomonas.PLoS One. 2012;7(8):e43118. doi: 10.1371/journal.pone.0043118. Epub 2012 Aug 10. PLoS One. 2012. PMID: 22900094 Free PMC article.
-
The intraflagellar transport machinery of Chlamydomonas reinhardtii.Traffic. 2003 Jul;4(7):435-42. doi: 10.1034/j.1600-0854.2003.t01-1-00103.x. Traffic. 2003. PMID: 12795688 Review.
-
Regulation of flagellar length in Chlamydomonas.Semin Cell Dev Biol. 2008 Dec;19(6):494-501. doi: 10.1016/j.semcdb.2008.07.005. Epub 2008 Jul 25. Semin Cell Dev Biol. 2008. PMID: 18692148 Free PMC article. Review.
Cited by
-
Non-ciliary Roles of IFT Proteins in Cell Division and Polycystic Kidney Diseases.Front Cell Dev Biol. 2020 Sep 22;8:578239. doi: 10.3389/fcell.2020.578239. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33072760 Free PMC article. Review.
-
New mutations in flagellar motors identified by whole genome sequencing in Chlamydomonas.Cilia. 2013 Oct 30;2(1):14. doi: 10.1186/2046-2530-2-14. Cilia. 2013. PMID: 24229452 Free PMC article.
-
Intraflagellar transport complex structure and cargo interactions.Cilia. 2013 Aug 14;2(1):10. doi: 10.1186/2046-2530-2-10. Cilia. 2013. PMID: 23945166 Free PMC article.
-
Chlamydomonas LZTFL1 mediates phototaxis via controlling BBSome recruitment to the basal body and its reassembly at the ciliary tip.Proc Natl Acad Sci U S A. 2021 Aug 31;118(35):e2101590118. doi: 10.1073/pnas.2101590118. Proc Natl Acad Sci U S A. 2021. PMID: 34446551 Free PMC article.
-
IFT27 links the BBSome to IFT for maintenance of the ciliary signaling compartment.Dev Cell. 2014 Nov 10;31(3):279-290. doi: 10.1016/j.devcel.2014.09.011. Epub 2014 Oct 30. Dev Cell. 2014. PMID: 25446516 Free PMC article.
References
-
- Wheatley D. N. (1995) Pathobiology 63, 222–238 - PubMed
-
- Bray D. (2001) Cell Movements: From Molecules to Motility, pp. 3–16, 225–241, Garland Publishing, New York, NY
-
- Praetorius H. A., Spring K. R. (2005) Annu. Rev. Physiol. 67, 515–529 - PubMed
-
- Davis E. E., Brueckner M., Katsanis N. (2006) Dev. Cell 11, 9–19 - PubMed
-
- Pazour G. J., Witman G. B. (2003) Curr. Opin. Cell Biol. 15, 105–110 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

