Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB
- PMID: 20436459
- PMCID: PMC3182123
- DOI: 10.1038/nature08966
Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB
Abstract
Polycomb group (PcG) proteins are transcriptional repressors that control processes ranging from the maintenance of cell fate decisions and stem cell pluripotency in animals to the control of flowering time in plants. In Drosophila, genetic studies identified more than 15 different PcG proteins that are required to repress homeotic (HOX) and other developmental regulator genes in cells where they must stay inactive. Biochemical analyses established that these PcG proteins exist in distinct multiprotein complexes that bind to and modify chromatin of target genes. Among those, Polycomb repressive complex 1 (PRC1) and the related dRing-associated factors (dRAF) complex contain an E3 ligase activity for monoubiquitination of histone H2A (refs 1-4). Here we show that the uncharacterized Drosophila PcG gene calypso encodes the ubiquitin carboxy-terminal hydrolase BAP1. Biochemically purified Calypso exists in a complex with the PcG protein ASX, and this complex, named Polycomb repressive deubiquitinase (PR-DUB), is bound at PcG target genes in Drosophila. Reconstituted recombinant Drosophila and human PR-DUB complexes remove monoubiquitin from H2A but not from H2B in nucleosomes. Drosophila mutants lacking PR-DUB show a strong increase in the levels of monoubiquitinated H2A. A mutation that disrupts the catalytic activity of Calypso, or absence of the ASX subunit abolishes H2A deubiquitination in vitro and HOX gene repression in vivo. Polycomb gene silencing may thus entail a dynamic balance between H2A ubiquitination by PRC1 and dRAF, and H2A deubiquitination by PR-DUB.
Figures

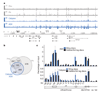
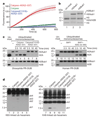
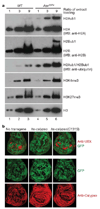
Comment in
-
The DUBle life of polycomb complexes.Dev Cell. 2010 Jun 15;18(6):878-80. doi: 10.1016/j.devcel.2010.06.001. Dev Cell. 2010. PMID: 20627069
References
-
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nature Rev. Genet. 2007;8:9–22. - PubMed
-
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. - PubMed
-
- Müller J, Verrijzer CP. Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr. Opin. Genet. Dev. 2009;19:150–158. - PubMed
-
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nature Rev. Mol. Cell Biol. 2009;10:697–708. - PubMed
-
- Pietersen AM, van Lohuizen M. Stem cell regulation by polycomb repressors: postponing commitment. Curr. Opin. Cell Biol. 2008;20:201–207. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

