Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs
- PMID: 20436462
- PMCID: PMC2868100
- DOI: 10.1038/nbt.1633
Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs
Erratum in
- Nat Biotechnol. 2010 Jul;28(7):756
Abstract
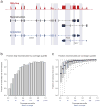
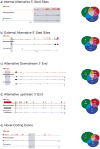
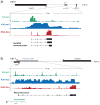
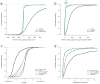
Massively parallel cDNA sequencing (RNA-Seq) provides an unbiased way to study a transcriptome, including both coding and noncoding genes. Until now, most RNA-Seq studies have depended crucially on existing annotations and thus focused on expression levels and variation in known transcripts. Here, we present Scripture, a method to reconstruct the transcriptome of a mammalian cell using only RNA-Seq reads and the genome sequence. We applied it to mouse embryonic stem cells, neuronal precursor cells and lung fibroblasts to accurately reconstruct the full-length gene structures for most known expressed genes. We identified substantial variation in protein coding genes, including thousands of novel 5' start sites, 3' ends and internal coding exons. We then determined the gene structures of more than a thousand large intergenic noncoding RNA (lincRNA) and antisense loci. Our results open the way to direct experimental manipulation of thousands of noncoding RNAs and demonstrate the power of ab initio reconstruction to render a comprehensive picture of mammalian transcriptomes.
Figures

Comment in
-
Advancing RNA-Seq analysis.Nat Biotechnol. 2010 May;28(5):421-3. doi: 10.1038/nbt0510-421. Nat Biotechnol. 2010. PMID: 20458303 No abstract available.
References
-
- Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. - PubMed
-
- Kapranov P, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science (New York, NY) 2007;316:1484–1488. 1138341 [pii] 10.1126/science.1138341. - PubMed
-
- Bertone P, et al. Global identification of human transcribed sequences with genome tiling arrays. Science (New York, NY) 2004;306:2242–2246. 1103388 [pii] 10.1126/science.1103388. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

