A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4
- PMID: 20436469
- PMCID: PMC2941216
- DOI: 10.1038/ng.580
A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4
Erratum in
- Nat Genet. 2010 Aug;42(8):727. Scott, James M [corrected to Scott, John M]
Abstract
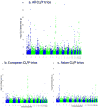
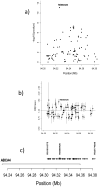
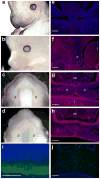
Case-parent trios were used in a genome-wide association study of cleft lip with and without cleft palate. SNPs near two genes not previously associated with cleft lip with and without cleft palate (MAFB, most significant SNP rs13041247, with odds ratio (OR) per minor allele = 0.704, 95% CI 0.635-0.778, P = 1.44 x 10(-11); and ABCA4, most significant SNP rs560426, with OR = 1.432, 95% CI 1.292-1.587, P = 5.01 x 10(-12)) and two previously identified regions (at chromosome 8q24 and IRF6) attained genome-wide significance. Stratifying trios into European and Asian ancestry groups revealed differences in statistical significance, although estimated effect sizes remained similar. Replication studies from several populations showed confirming evidence, with families of European ancestry giving stronger evidence for markers in 8q24, whereas Asian families showed stronger evidence for association with MAFB and ABCA4. Expression studies support a role for MAFB in palatal development.
Figures

References
-
- Mossey PA, Little J, Munger RG, Dixon MJ, Shaw WC. Cleft lip and palate. Lancet. 2009;374:1773–1785. - PubMed
-
- Jugessur A, Farlie PG, Kilpatrick N. The genetics of isolated orofacial clefts: from genotypes to subphenotypes. Oral Diseases. 2009;15:437–453. - PubMed
-
- Mitchell LE. Twin studies in oral cleft research. In: Wyszynski DF, editor. Cleft Lip and Palate. Oxford University Press; New York: 2002. pp. 214–221.
Publication types
MeSH terms
Substances
Grants and funding
- R37 DE008559/DE/NIDCR NIH HHS/United States
- JK02-AEO15291/PHS HHS/United States
- P50-DE-16215/DE/NIDCR NIH HHS/United States
- R21-DE016930/DE/NIDCR NIH HHS/United States
- R01 DE016148/DE/NIDCR NIH HHS/United States
- R01-DE016877/DE/NIDCR NIH HHS/United States
- R01 DE012472/DE/NIDCR NIH HHS/United States
- R01-DE014677/DE/NIDCR NIH HHS/United States
- R01-DE09886/DE/NIDCR NIH HHS/United States
- R21 DE016930/DE/NIDCR NIH HHS/United States
- R01 DE009886/DE/NIDCR NIH HHS/United States
- R01-HD390661/HD/NICHD NIH HHS/United States
- R01-DE016148/DE/NIDCR NIH HHS/United States
- R37-DE08559/DE/NIDCR NIH HHS/United States
- U01-DE-018993/DE/NIDCR NIH HHS/United States
- U01 DE018993/DE/NIDCR NIH HHS/United States
- MT-11263/CAPMC/ CIHR/Canada
- R03 AR055313/AR/NIAMS NIH HHS/United States
- R01-DE012472/DE/NIDCR NIH HHS/United States
- R01 DE014581/DE/NIDCR NIH HHS/United States
- HHSN268200782096C/HG/NHGRI NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- R00 DE018413/DE/NIDCR NIH HHS/United States
- R01-DE-014581/DE/NIDCR NIH HHS/United States
- R21 DE016877/DE/NIDCR NIH HHS/United States
- P50 DE016215/DE/NIDCR NIH HHS/United States
- P50-DE016215/DE/NIDCR NIH HHS/United States
- K99 DE018413/DE/NIDCR NIH HHS/United States
- K99-DE-018441/DE/NIDCR NIH HHS/United States
- R03-AR-055313/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

