Quantitative phosphoproteomic analysis of T cell receptor signaling in diabetes prone and resistant mice
- PMID: 20438120
- PMCID: PMC3603002
- DOI: 10.1021/pr100035b
Quantitative phosphoproteomic analysis of T cell receptor signaling in diabetes prone and resistant mice
Abstract
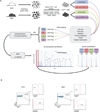
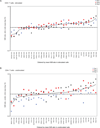
Type 1 diabetes, in human patients and NOD mice, results from an immune attack on insulin-producing beta-cells of the pancreas by autoreactive T lymphocytes. In NOD mice, genetically controlled perturbations in the signaling pathways downstream of the antigen-specific T cell receptor (TCR) may be instrumental in the altered responses of T cells, manifest as inefficient induction of apoptosis after recognition of self-antigens in the thymus or as perturbed reactivity of mature T cells in peripheral organs. To map this signaling difference(s), we have used mass spectrometry-based quantitative phosphoproteomics to compare the activation of primary CD4(+) T cells of diabetes-prone NOD and -resistant B6.H2g7 mice. Immunoprecipitation and IMAC purification of tyrosine-phosphorylated peptides, combined with a stable-isotope iTRAQ labeling, enabled us to identify and quantify over 77 phosphorylation events in 54 different proteins downstream of TCR stimulation of primary CD4(+) T cells. This analysis showed a generally higher level of phosphotyrosine in activated NOD cells, as well as several phosphorylation sites that appeared to be differentially regulated in these two strains (involving TXK, CD5, PAG1, and ZAP-70). These data highlight the differences in signaling between CD4(+) T cell compartments of NOD and B6g7 mice and may underlie the dysregulation of T cells in NOD mice.
Figures




References
-
- Kikutani H, Makino S. The murine autoimmune diabetes model: NOD and related strains. Adv. Immunol. 1992;51:285–322. - PubMed
-
- Concannon P, Rich SS, Nepom GT. Genetics of type 1A diabetes. N. Engl. J. Med. 2009;360(16):1646–1654. - PubMed
-
- Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, Plagnol V, Pociot F, Schuilenburg H, Smyth DJ, Stevens H, Todd JA, Walker NM, Rich SS. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009 - PMC - PubMed
-
- Bluestone JA, Tang Q, Sedwick CE. T Regulatory Cells in Autoimmune Diabetes: Past Challenges, Future Prospects. J Clin Immunol. 2008 - PubMed
-
- Kishimoto H, Sprent J. A defect in central tolerance in NOD mice. Nat. Immunol. 2001;2(11):1025–1031. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

