Creating PWMs of transcription factors using 3D structure-based computation of protein-DNA free binding energies
- PMID: 20438625
- PMCID: PMC2879287
- DOI: 10.1186/1471-2105-11-225
Creating PWMs of transcription factors using 3D structure-based computation of protein-DNA free binding energies
Abstract
Background: Knowledge of transcription factor-DNA binding patterns is crucial for understanding gene transcription. Numerous DNA-binding proteins are annotated as transcription factors in the literature, however, for many of them the corresponding DNA-binding motifs remain uncharacterized.
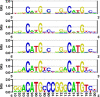
Results: The position weight matrices (PWMs) of transcription factors from different structural classes have been determined using a knowledge-based statistical potential. The scoring function calibrated against crystallographic data on protein-DNA contacts recovered PWMs of various members of widely studied transcription factor families such as p53 and NF-kappaB. Where it was possible, extensive comparison to experimental binding affinity data and other physical models was made. Although the p50p50, p50RelB, and p50p65 dimers belong to the same family, particular differences in their PWMs were detected, thereby suggesting possibly different in vivo binding modes. The PWMs of p63 and p73 were computed on the basis of homology modeling and their performance was studied using upstream sequences of 85 p53/p73-regulated human genes. Interestingly, about half of the p63 and p73 hits reported by the Match algorithm in the altogether 126 promoters lay more than 2 kb upstream of the corresponding transcription start sites, which deviates from the common assumption that most regulatory sites are located more proximal to the TSS. The fact that in most of the cases the binding sites of p63 and p73 did not overlap with the p53 sites suggests that p63 and p73 could influence the p53 transcriptional activity cooperatively. The newly computed p50p50 PWM recovered 5 more experimental binding sites than the corresponding TRANSFAC matrix, while both PWMs showed comparable receiver operator characteristics.
Conclusions: A novel algorithm was developed to calculate position weight matrices from protein-DNA complex structures. The proposed algorithm was extensively validated against experimental data. The method was further combined with Homology Modeling to obtain PWMs of factors for which crystallographic complexes with DNA are not yet available. The performance of PWMs obtained in this work in comparison to traditionally constructed matrices demonstrates that the structure-based approach presents a promising alternative to experimental determination of transcription factor binding properties.
Figures





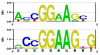


References
-
- Liu LA, Bader JS. Ab initio prediction of transcription factor binding sites. Pacific Symposium on Biocomputing. 2007;12:484–495. full_text. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

