Structure of the Newcastle disease virus F protein in the post-fusion conformation
- PMID: 20439109
- PMCID: PMC2877518
- DOI: 10.1016/j.virol.2010.03.050
Structure of the Newcastle disease virus F protein in the post-fusion conformation
Abstract
The paramyxovirus F protein is a class I viral membrane fusion protein which undergoes a significant refolding transition during virus entry. Previous studies of the Newcastle disease virus, human parainfluenza virus 3 and parainfluenza virus 5 F proteins revealed differences in the pre- and post-fusion structures. The NDV Queensland (Q) F structure lacked structural elements observed in the other two structures, which are key to the refolding and fusogenic activity of F. Here we present the NDV Australia-Victoria (AV) F protein post-fusion structure and provide EM evidence for its folding to a pre-fusion form. The NDV AV F structure contains heptad repeat elements missing in the previous NDV Q F structure, forming a post-fusion six-helix bundle (6HB) similar to the post-fusion hPIV3 F structure. Electrostatic and temperature factor analysis of the F structures points to regions of these proteins that may be functionally important in their membrane fusion activity.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures


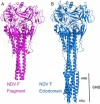
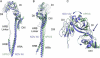


Similar articles
-
Crystallization and preliminary X-ray diffraction analysis of post-fusion six-helix bundle core structure from Newcastle disease virus F protein.Acta Crystallogr D Biol Crystallogr. 2003 Jul;59(Pt 7):1296-8. doi: 10.1107/s0907444903009909. Epub 2003 Jun 27. Acta Crystallogr D Biol Crystallogr. 2003. PMID: 12832792
-
Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation.Nature. 2006 Jan 5;439(7072):38-44. doi: 10.1038/nature04322. Nature. 2006. PMID: 16397490 Free PMC article.
-
Six-helix bundle assembly and characterization of heptad repeat regions from the F protein of Newcastle disease virus.J Gen Virol. 2002 Mar;83(Pt 3):623-629. doi: 10.1099/0022-1317-83-3-623. J Gen Virol. 2002. PMID: 11842257
-
Structural basis of viral invasion: lessons from paramyxovirus F.Curr Opin Struct Biol. 2007 Aug;17(4):427-36. doi: 10.1016/j.sbi.2007.08.016. Epub 2007 Sep 17. Curr Opin Struct Biol. 2007. PMID: 17870467 Free PMC article. Review.
-
Structure and function of a paramyxovirus fusion protein.Biochim Biophys Acta. 2003 Jul 11;1614(1):73-84. doi: 10.1016/s0005-2736(03)00164-0. Biochim Biophys Acta. 2003. PMID: 12873767 Review.
Cited by
-
Electron tomography imaging of surface glycoproteins on human parainfluenza virus 3: association of receptor binding and fusion proteins before receptor engagement.mBio. 2015 Feb 17;6(1):e02393-14. doi: 10.1128/mBio.02393-14. mBio. 2015. PMID: 25691596 Free PMC article.
-
Pigeon paramyxovirus type 1 variants with polybasic F protein cleavage site but strikingly different pathogenicity.Virus Genes. 2014 Dec;49(3):502-6. doi: 10.1007/s11262-014-1111-7. Epub 2014 Sep 17. Virus Genes. 2014. PMID: 25228150
-
Structure and organization of paramyxovirus particles.Curr Opin Virol. 2017 Jun;24:105-114. doi: 10.1016/j.coviro.2017.05.004. Epub 2017 Jun 8. Curr Opin Virol. 2017. PMID: 28601688 Free PMC article. Review.
-
The paramyxovirus fusion protein C-terminal region: mutagenesis indicates an indivisible protein unit.J Virol. 2012 Mar;86(5):2600-9. doi: 10.1128/JVI.06546-11. Epub 2011 Dec 14. J Virol. 2012. PMID: 22171273 Free PMC article.
-
Emergence and adaptive evolution of Nipah virus.Transbound Emerg Dis. 2020 Jan;67(1):121-132. doi: 10.1111/tbed.13330. Epub 2019 Sep 10. Transbound Emerg Dis. 2020. PMID: 31408582 Free PMC article.
References
-
- Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. - PubMed
-
- Baker K, et al. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell. 1999;3:309–319. - PubMed
-
- Bullough PA, et al. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. - PubMed
-
- Calder LJ, et al. Electron microscopy of the human respiratory syncytial virus fusion protein and complexes that it forms with monoclonal antibodies. Virology. 2000;271:122–131. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

