A low-polynomial algorithm for assembling clusters of orthologous groups from intergenomic symmetric best matches
- PMID: 20439257
- PMCID: PMC2881409
- DOI: 10.1093/bioinformatics/btq229
A low-polynomial algorithm for assembling clusters of orthologous groups from intergenomic symmetric best matches
Abstract
Motivation: Identifying orthologous genes in multiple genomes is a fundamental task in comparative genomics. Construction of intergenomic symmetrical best matches (SymBets) and joining them into clusters is a popular method of ortholog definition, embodied in several software programs. Despite their wide use, the computational complexity of these programs has not been thoroughly examined.
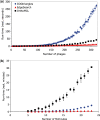
Results: In this work, we show that in the standard approach of iteration through all triangles of SymBets, the memory scales with at least the number of these triangles, O(g(3)) (where g = number of genomes), and construction time scales with the iteration through each pair, i.e. O(g(6)). We propose the EdgeSearch algorithm that iterates over edges in the SymBet graph rather than triangles of SymBets, and as a result has a worst-case complexity of only O(g(3)log g). Several optimizations reduce the run-time even further in realistically sparse graphs. In two real-world datasets of genomes from bacteriophages (POGs) and Mollicutes (MOGs), an implementation of the EdgeSearch algorithm runs about an order of magnitude faster than the original algorithm and scales much better with increasing number of genomes, with only minor differences in the final results, and up to 60 times faster than the popular OrthoMCL program with a 90% overlap between the identified groups of orthologs.
Availability and implementation: C++ source code freely available for download at ftp.ncbi.nih.gov/pub/wolf/COGs/COGsoft/.
Supplementary information: Supplementary materials are available at Bioinformatics online.
Figures


Similar articles
-
JustOrthologs: a fast, accurate and user-friendly ortholog identification algorithm.Bioinformatics. 2019 Feb 15;35(4):546-552. doi: 10.1093/bioinformatics/bty669. Bioinformatics. 2019. PMID: 30084941 Free PMC article.
-
Clusters of orthologous genes for 41 archaeal genomes and implications for evolutionary genomics of archaea.Biol Direct. 2007 Nov 27;2:33. doi: 10.1186/1745-6150-2-33. Biol Direct. 2007. PMID: 18042280 Free PMC article.
-
Ortholog clustering on a multipartite graph.IEEE/ACM Trans Comput Biol Bioinform. 2007 Jan-Mar;4(1):17-27. doi: 10.1109/TCBB.2007.1004. IEEE/ACM Trans Comput Biol Bioinform. 2007. PMID: 17277410
-
TwoPaCo: an efficient algorithm to build the compacted de Bruijn graph from many complete genomes.Bioinformatics. 2017 Dec 15;33(24):4024-4032. doi: 10.1093/bioinformatics/btw609. Bioinformatics. 2017. PMID: 27659452
-
Automatic clustering of orthologs and in-paralogs from pairwise species comparisons.J Mol Biol. 2001 Dec 14;314(5):1041-52. doi: 10.1006/jmbi.2000.5197. J Mol Biol. 2001. PMID: 11743721
Cited by
-
Novel Bacillus-Infecting Bacteriophage B13-The Founding Member of the Proposed New Genus Bunatrivirus.Viruses. 2022 Oct 19;14(10):2300. doi: 10.3390/v14102300. Viruses. 2022. PMID: 36298855 Free PMC article.
-
Scandinavium goeteborgense gen. nov., sp. nov., a New Member of the Family Enterobacteriaceae Isolated From a Wound Infection, Carries a Novel Quinolone Resistance Gene Variant.Front Microbiol. 2019 Nov 5;10:2511. doi: 10.3389/fmicb.2019.02511. eCollection 2019. Front Microbiol. 2019. PMID: 31781055 Free PMC article.
-
Identification of sumoylated proteins in the silkworm Bombyx mori.Int J Mol Sci. 2014 Dec 1;15(12):22011-27. doi: 10.3390/ijms151222011. Int J Mol Sci. 2014. PMID: 25470021 Free PMC article.
-
Informative Regions In Viral Genomes.Viruses. 2021 Jun 18;13(6):1164. doi: 10.3390/v13061164. Viruses. 2021. PMID: 34207030 Free PMC article.
-
Inferring Orthologs: Open Questions and Perspectives.Genomics Insights. 2016 Feb 25;9:17-28. doi: 10.4137/GEI.S37925. eCollection 2016. Genomics Insights. 2016. PMID: 26966373 Free PMC article. Review.
References
-
- Alexeyenko A, et al. Automatic clustering of orthologs and inparalogs shared by multiple proteomes. Bioinformatics. 2006;22:e9–e15. - PubMed
-
- Brenner SE. Errors in genome annotation. Trends Genet. 1999;15:132–133. - PubMed
-
- Deluca TF, et al. Roundup: a multi-genome repository of orthologs and evolutionary distances. Bioinformatics. 2006;22:2044–2046. - PubMed

