Characterization of the role of the pathogenicity island and vapG in the virulence of the intracellular actinomycete pathogen Rhodococcus equi
- PMID: 20439471
- PMCID: PMC2916281
- DOI: 10.1128/IAI.00081-10
Characterization of the role of the pathogenicity island and vapG in the virulence of the intracellular actinomycete pathogen Rhodococcus equi
Abstract
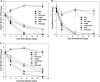
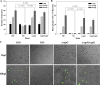
Rhodococcus equi, a facultative intracellular pathogen of macrophages, causes severe, life-threatening pneumonia in young foals and in people with underlying immune deficiencies. R. equi virulence is dependent on the presence of a large virulence plasmid that houses a pathogenicity island (PAI) encoding a novel family of surface-localized and secreted proteins of largely unknown function termed the virulence-associated proteins (VapACDEFGHI). To date, vapA and its positive regulators virR and orf8 are the only experimentally established virulence genes residing on the virulence plasmid. In this study, a PAI deletion mutant was constructed and, as anticipated, was attenuated for growth both in macrophages and in mice due to the absence of vapA expression. Expression of vapA in the PAI mutant from a constitutive promoter, thereby eliminating the requirement for the PAI-encoded vapA regulators, resulted in delayed bacterial clearance in vivo, yet full virulence was not restored, indicating that additional virulence genes are indeed located within the deleted pathogenicity island region. Based on previous reports demonstrating that the PAI-carried gene vapG is highly upregulated in macrophages and in the lungs of R. equi-infected foals, we hypothesized that vapG could be an important virulence factor. However, analysis of a marked vapG deletion mutant determined the gene to be dispensable for growth in macrophages and in vivo in mice.
Figures

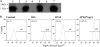


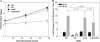
Similar articles
-
Deletion of vapA encoding Virulence Associated Protein A attenuates the intracellular actinomycete Rhodococcus equi.Mol Microbiol. 2003 Oct;50(1):115-28. doi: 10.1046/j.1365-2958.2003.03689.x. Mol Microbiol. 2003. PMID: 14507368
-
Role of the 85-kilobase plasmid and plasmid-encoded virulence-associated protein A in intracellular survival and virulence of Rhodococcus equi.Infect Immun. 1999 Jul;67(7):3548-57. doi: 10.1128/IAI.67.7.3548-3557.1999. Infect Immun. 1999. PMID: 10377138 Free PMC article.
-
Identification of a VapA virulence factor functional homolog in Rhodococcus equi isolates housing the pVAPB plasmid.PLoS One. 2018 Oct 4;13(10):e0204475. doi: 10.1371/journal.pone.0204475. eCollection 2018. PLoS One. 2018. PMID: 30286098 Free PMC article.
-
Molecular and infection biology of the horse pathogen Rhodococcus equi.FEMS Microbiol Rev. 2009 Sep;33(5):870-91. doi: 10.1111/j.1574-6976.2009.00181.x. Epub 2009 Apr 23. FEMS Microbiol Rev. 2009. PMID: 19453748 Review.
-
Pathogenesis and virulence of Rhodococcus equi.Vet Microbiol. 1997 Jun 16;56(3-4):257-68. doi: 10.1016/s0378-1135(97)00094-1. Vet Microbiol. 1997. PMID: 9226840 Review.
Cited by
-
The steroid catabolic pathway of the intracellular pathogen Rhodococcus equi is important for pathogenesis and a target for vaccine development.PLoS Pathog. 2011 Aug;7(8):e1002181. doi: 10.1371/journal.ppat.1002181. Epub 2011 Aug 25. PLoS Pathog. 2011. PMID: 21901092 Free PMC article.
-
Structural characterisation of the virulence-associated protein VapG from the horse pathogen Rhodococcus equi.Vet Microbiol. 2015 Aug 31;179(1-2):42-52. doi: 10.1016/j.vetmic.2015.01.027. Epub 2015 Feb 9. Vet Microbiol. 2015. PMID: 25746683 Free PMC article.
-
Rhodococcus parequi sp. nov., a new species isolated from equine farm soil closely related to the pathogen Rhodococcus equi.Int J Syst Evol Microbiol. 2025 Mar;75(3):006679. doi: 10.1099/ijsem.0.006679. Int J Syst Evol Microbiol. 2025. PMID: 40063668 Free PMC article.
-
The N-terminal domain is required for cell surface localisation of VapA, a member of the Vap family of Rhodococcus equi virulence proteins.PLoS One. 2024 Feb 29;19(2):e0298900. doi: 10.1371/journal.pone.0298900. eCollection 2024. PLoS One. 2024. PMID: 38421980 Free PMC article.
-
Molecular analysis of the chromosomal 16S rRNA gene and vapA plasmid gene of Polish field strains of R. equi.PLoS One. 2018 Sep 25;13(9):e0204024. doi: 10.1371/journal.pone.0204024. eCollection 2018. PLoS One. 2018. PMID: 30252885 Free PMC article.
References
-
- Ainsworth, D. M., S. W. Eicker, A. E. Yeagar, C. R. Sweeney, L. Viel, D. Tesarowski, J. P. Lavoie, A. Hoffman, M. R. Paradis, S. M. Reed, H. N. Erb, E. Davidow, and M. Nalevanko. 1998. Associations between physical examination, laboratory, and radiographic findings and outcome and subsequent racing performance of foals with Rhodococcus equi infection: 115 cases (1984-1992). J. Am. Vet. Med. Assoc. 213:510-515. - PubMed
-
- Arlotti, M., G. Zoboli, G. L. Moscatelli, G. Magnani, R. Maserati, V. Borghi, M. Andreoni, M. Libanore, L. Bonazzi, A. Piscina, and R. Ciammarughi. 1996. Rhodococcus equi infection in HIV-positive subjects: a retrospective analysis of 24 cases. Scand. J. Infect. Dis. 28:463-467. - PubMed
-
- Benoit, S., A. Benachour, S. Taouji, Y. Auffray, and A. Hartke. 2001. Induction of vap genes encoded by the virulence plasmid of Rhodococcus equi during acid tolerance response. Res. Microbiol. 152:439-449. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

