Peptide antagonism as a mechanism for NK cell activation
- PMID: 20439706
- PMCID: PMC2890497
- DOI: 10.1073/pnas.0913745107
Peptide antagonism as a mechanism for NK cell activation
Abstract
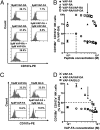
Inhibition of natural killer (NK) cells is mediated by MHC class I receptors including the killer cell Ig-like receptor (KIR). We demonstrate that HLA-C binding peptides can function as altered peptide ligands for KIR and antagonize the inhibition mediated by KIR2DL2/KIR2DL3. Antagonistic peptides promote clustering of KIR at the interface of effector and target cells, but do not result in inhibition of NK cells. Our data show that, as for T cells, small changes in the peptide content of MHC class I can regulate NK cell activity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

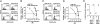
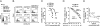

Comment in
-
Antagonizing inhibition gets NK cells going.Proc Natl Acad Sci U S A. 2010 Jun 8;107(23):10333-4. doi: 10.1073/pnas.1005636107. Epub 2010 Jun 2. Proc Natl Acad Sci U S A. 2010. PMID: 20534579 Free PMC article. No abstract available.
References
-
- Malnati MS, et al. Peptide specificity in the recognition of MHC class I by natural killer cell clones. Science. 1995;267:1016–1018. - PubMed
-
- Maenaka K, et al. Killer cell immunoglobulin receptors and T cell receptors bind peptide-major histocompatibility complex class I with distinct thermodynamic and kinetic properties. J Biol Chem. 1999;274:28329–28334. - PubMed
-
- Thananchai H, et al. Cutting Edge: Allele-specific and peptide-dependent interactions between KIR3DL1 and HLA-A and HLA-B. J Immunol. 2007;178:33–37. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

