Cotranslational structure acquisition of nascent polypeptides monitored by NMR spectroscopy
- PMID: 20439768
- PMCID: PMC2889043
- DOI: 10.1073/pnas.0914300107
Cotranslational structure acquisition of nascent polypeptides monitored by NMR spectroscopy
Abstract
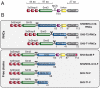
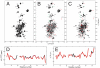
The folding of proteins in living cells may start during their synthesis when the polypeptides emerge gradually at the ribosomal exit tunnel. However, our current understanding of cotranslational folding processes at the atomic level is limited. We employed NMR spectroscopy to monitor the conformation of the SH3 domain from alpha-spectrin at sequential stages of elongation via in vivo ribosome-arrested (15)N,(13)C-labeled nascent polypeptides. These nascent chains exposed either the entire SH3 domain or C-terminally truncated segments thereof, thus providing snapshots of the translation process. We show that nascent SH3 polypeptides remain unstructured during elongation but fold into a compact, native-like beta-sheet assembly when the entire sequence information is available. Moreover, the ribosome neither imposes major conformational constraints nor significantly interacts with exposed unfolded nascent SH3 domain moieties. Our data provide evidence for a domainwise folding of the SH3 domain on ribosomes without significant population of folding intermediates. The domain follows a thermodynamically favorable pathway in which sequential folding units are stabilized, thus avoiding kinetic traps during the process of cotranslational folding.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Ban N, et al. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. - PubMed
-
- Yusupov MM, et al. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. - PubMed
-
- Ramakrishnan V. Ribosome structure and the mechanism of translation. Cell. 2002;108:557–572. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

