Functional genomics analysis of the Saccharomyces cerevisiae iron responsive transcription factor Aft1 reveals iron-independent functions
- PMID: 20439772
- PMCID: PMC2900968
- DOI: 10.1534/genetics.110.117531
Functional genomics analysis of the Saccharomyces cerevisiae iron responsive transcription factor Aft1 reveals iron-independent functions
Abstract
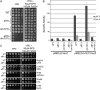
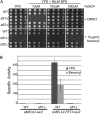
The Saccharomyces cerevisiae transcription factor Aft1 is activated in iron-deficient cells to induce the expression of iron regulon genes, which coordinate the increase of iron uptake and remodel cellular metabolism to survive low-iron conditions. In addition, Aft1 has been implicated in numerous cellular processes including cell-cycle progression and chromosome stability; however, it is unclear if all cellular effects of Aft1 are mediated through iron homeostasis. To further investigate the cellular processes affected by Aft1, we identified >70 deletion mutants that are sensitive to perturbations in AFT1 levels using genome-wide synthetic lethal and synthetic dosage lethal screens. Our genetic network reveals that Aft1 affects a diverse range of cellular processes, including the RIM101 pH pathway, cell-wall stability, DNA damage, protein transport, chromosome stability, and mitochondrial function. Surprisingly, only a subset of mutants identified are sensitive to extracellular iron fluctuations or display genetic interactions with mutants of iron regulon genes AFT2 or FET3. We demonstrate that Aft1 works in parallel with the RIM101 pH pathway and the role of Aft1 in DNA damage repair is mediated by iron. In contrast, through both directed studies and microarray transcriptional profiling, we show that the role of Aft1 in chromosome maintenance and benomyl resistance is independent of its iron regulatory role, potentially through a nontranscriptional mechanism.
Figures




References
-
- Abelson, J. N., M.I. Simon, C. Guthrie and G.R. Fink, 2004. Guide to Yeast Genetics and Molecular Biology. Elsevier Academic Press, San Diego, CA.
-
- Baetz, K., V. Measday and B. Andrews, 2006. Revealing hidden relationships among yeast genes involved in chromosome segregation using systematic synthetic lethal and synthetic dosage lethal screens. Cell Cycle 5 592–595. - PubMed
-
- Bennett, C. B., L. K. Lewis, G. Karthikeyan, K. S. Lobachev, Y. H. Jin et al., 2001. Genes required for ionizing radiation resistance in yeast. Nat. Genet. 29 426–434. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

