How antibiotics kill bacteria: from targets to networks
- PMID: 20440275
- PMCID: PMC2896384
- DOI: 10.1038/nrmicro2333
How antibiotics kill bacteria: from targets to networks
Abstract
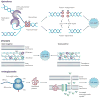
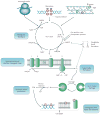
Antibiotic drug-target interactions, and their respective direct effects, are generally well characterized. By contrast, the bacterial responses to antibiotic drug treatments that contribute to cell death are not as well understood and have proven to be complex as they involve many genetic and biochemical pathways. In this Review, we discuss the multilayered effects of drug-target interactions, including the essential cellular processes that are inhibited by bactericidal antibiotics and the associated cellular response mechanisms that contribute to killing. We also discuss new insights into these mechanisms that have been revealed through the study of biological networks, and describe how these insights, together with related developments in synthetic biology, could be exploited to create new antibacterial therapies.
Figures


References
-
- Walsh C. Antibiotics: actions, origins, resistance. ASM Press; Washington, D.C: 2003.
-
- Fleming A. On antibacterial action of culture of penicillium, with special reference to their use in isolation of B. influenzae. British Journal of Experimental Pathology. 1929;10:226–236. - PubMed
-
- Taubes G. The bacteria fight back. Science. 2008;321:356–61. - PubMed
-
- Floss HG, Yu TW. Rifamycin-mode of action, resistance, and biosynthesis. Chem Rev. 2005;105:621–32. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

